Difference between revisions of "ExpressionView"
| Line 32: | Line 32: | ||
= Additional documentation = | = Additional documentation = | ||
| − | * [[Media:Expressionview.ordering|Ordering algorithm]] | + | * [[Media:Expressionview.ordering.pdf|Ordering algorithm]] |
| + | * {{Pdf|Expressionview.ordering.pdf|Ordering algorithm}} | ||
* [http://maya:7575/ExpressionView/doc ActionScript source code documentation] | * [http://maya:7575/ExpressionView/doc ActionScript source code documentation] | ||
* [[Media:Expressionview-xml|XML file structure]] | * [[Media:Expressionview-xml|XML file structure]] | ||
Revision as of 10:44, 16 November 2009
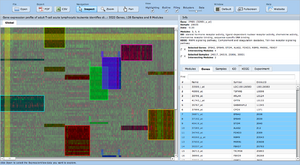
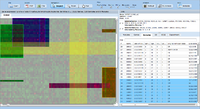
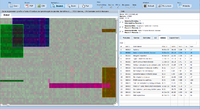
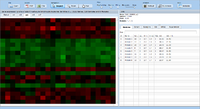
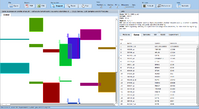
ExpressionView is an R package that provides an interactive environment to explore biclusters identified in gene expression data. A sophisticated ordering algorithm is used to present the biclusters in a visually appealing layout that provides an intuitive summary of the results. From this overview, the user can select individual biclusters and access all the biologically relevant data associated with it. The package is aimed to facilitate the collaboration between bioinformaticians and life scientists who are not familiar with the R language.
Contents
Launch
- Launch ExpressionView
- Launch ExpressionView with ALL data (8 modules)
- Launch ExpressionView with ALL data (108 modules)
Screenshots
Download
Tutorials
There are several tutorials describing how to use ExpressionView: