How genetic alterations propagate to organismal level traits
Léonard Jequier, Cynthia Meizoso, Dariush Mollet
Supervisor : Sarvenaz Choobdar
Introduction
Can you predict a cardiovascular disease from a certain gene expression level ?
In our project, we tried to link groups of similarly expressed genes to GWAS files in order to determine whether they were related to a phenotype. It is quite a challenge since it is new to link expression data to phenotypes from external data and with the help of bioinformatical tools (R Studio with the packages isa2, pascal and also a gene ontology).
Methods Our raw data was a set of protein encoding genes expression (555 genes from more than 19'000 healthy, randomized individuals) obtained from CoLaus. We also had the corresponding ensembl gene IDs. First, the plan was to do a biclustering on the expressions with ISA ([1] Iterative Signature Algorithm) and then map those groups (which are called modules) to a GWAS set, using Pascal ([2] Pathway Scoring Algorithm), in order to link them to phenotypes.
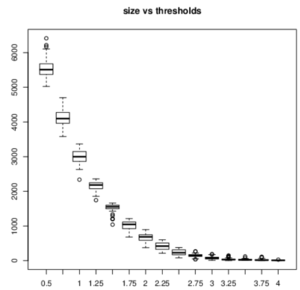
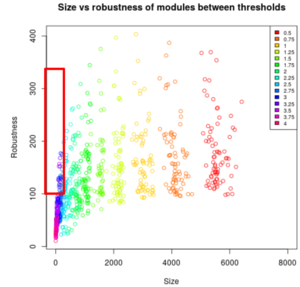
ISA is a biclustering tool that helped us aggregate the genes that are similarly expressed. Depending on the threshold, it finds the expression levels that are too far from the mean and combine a score of 0; if they are close to the mean, it will combine a score that is bigger than 0. We first had to normalize the data (log transformation) and then repeat the ISA to choose the threshold and module size. The threshold means "how far you are from gene expression", and small modules have really similar genes. We first needed to choose a size and threshold and then pick the best robustness of them. The robustness is proportional to the number of times the same module came out for each iteration.
Our next questions were: why do isa modules contain similarly expressed genes, and are these differences genetically associated to a phen? We used 16 GWASs from the Cardiovascular Disease set (p-values for SNP-phenotype associations). Pascal aggregates these p-values to our ISA modules to create disease modules. The output is a table with, for each GWAS that was used, a number of modules tested, a number of modules that came out significant, and the p-value associated to the link GWAS-module. Because of multiple hypotheses testing, a correction of the p-values was required.
Furthemore, a Gene Ontolofy enrichment (GO enrichment, [3] David Bioinformatical Database) was used to better understand the biological process. If, for a certain gene related to a certain phenotype, the gene expression is higher, then we call this phenomena an enrichment.
Results The biclustering gave us some trouble because it is important to find the right threshold and the right module size, which is a very arbitrary decision. If we had had more time, we would have run ISA several other times in order to be more sure of what is considered as a good signal.
The smaller the threshold is, the bigger is the size of the modules meaning that a lot of genes are in a module and that the biclustering was less restrictive. In our case, we needed the smaller size as possible, but not too much otherwise the robustness was too small. We got many modules and only a few of them had the right parameters (see red rectangle on the image below).
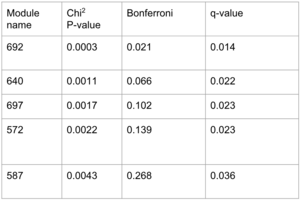
The obtained p-values from Pascal represent the significance of the association of the modules to one GWAS. Taking a look at the q-value rather than the adjusted p-values from Bonferroni correction, we found five signicant disease modules.
We compared our p-values to what we would have obtained if random (qqplot). It looks like more than five modules give a good signal for the GWAS on coronary-artery diseases, as well as for all the GWASes but less great.

We decided to do the GO enrichment on the five disease modules, but almost none gave a conclusive result. Only one disease module indeed gave a significant p-value for its association to the GTP binding, which is quite a vast molecular function.
Conclusion
In overall, it is difficult not only to get biological information, but relevant as well, from available repositories. It demands a good amount of time, and you may easily make errors, especially if you are not aware enough of the programs and softwares you use.
One should probably check the tools before using them. What tests are done ? What are the null hypothese ? We lost a a considerable amount of time trying to answer these questions. Also, we didn’t really discuss design experiment or statistical power. We probably should have asked ourselves "which steps are useful and why ?" before going straight into the project. What does each step prove ? What steps are crucial, or not ?
We actually did not give the GWASs a critical look before using them which allows us to only make poor interpretation. The software R is maybe not suited for this kind of project. Packages are well done, nevertheless working with object oriented tools would be better, because R is not strict at all.
Cheerful thanks to our supervisor for patiently taking time answer our questions and show us the right way many, many times throughout the project.