Modelling pathways crosstalks as closed walks and cycles on graphs
Contents
Modelling pathways crosstalk in gene/protein networks as closed walks & cycles on graphs
Definitions
Our project was to analyse and quantify interactions between different biological pathways. Those interactions are named closed walks. A closed walks is a sequence of interactions between genes that starts and ends at the same gene. A cycle is a closed walk that never goes twice through the same gene except for the gene from which the interactions start and end. A cross talk is a closed walk that goes through genes from two different pathways, meaning that those pathways communicate and regulate each other through their genes/proteins interactions.
Research questions
We had 3 research questions:
- Are some functional pathways more prone to crosstalk than the others? If so, which ones?
- What are the genes that are apparent “entry points” for the crosstalk with other pathways?
- Is crosstalk a symmetric process, i.e. do both pathways “need each other” the same, or is one rather “exploiting” the other for input stimuli?
the analysis concentrates on closed walks of length 2, 3 and 4 only, for computational power reasons.
Methods
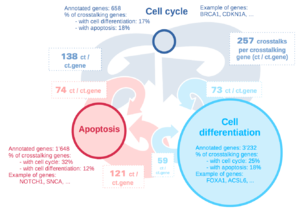
To answer the research questions we were provided with data containing genes' names, for each gene was indicated the different biological pathways it played a role into (each gene is involved in many different pathways). We first selected three different pathways to analyse, so that we could compare the interactions between each pair of those pathways. The chosen pathways were:
- Cell differentiation
- Mitotic cell cycle
- Programmed cell death
We also got a csv file containing all the interactions between those genes. This file is made of the name of the source gene and the name of the target gene and a sign: -1 or 1 that indicates if the interaction is an activation or an inhibition. Each of these three directed graphs was converted into an adjacency matrix. Each adjacency matrix was raised to the power of 2, 3 and 4.
To extract information from this data we used NetworkX on python. For each pathway we extracted a list of all the genes involved in it. Then for each possible pair of the three pathways we deleted the genes that belonged to the intersection. The genes in the intersection participate in both pathways so if they communicate we can't know if this is crosstalk. For each pair of pathway we have two unique lists of genes with no common genes between them. For each of those lists we built a directed graph representing the interactions between the genes of each pathways. Since the graph is directed it contains the information saying which gene initiate the interaction and which gene receive it. We then built a directed graph with the two lists of genes fused to represent the interactions between the two pathways. So for a pair of pathways to analyse we have:
- one directed graph per pathway
- one directed graph containing genes from both pathways
Since we are interested in closed walks of length 2, 3 and 4 we extract the diagonal of those matrices. Those diagonals tell us for each gene, in how many closed walks it is involved. If there is a closed walks of length 2 between gene A and gene B, this closed walk could start at gene A, go to B and come back to A: 2 interactions, length =2. But it is also true that this closed walk starts at gene B then goes to A and back to B. So the diagonal matrix will associate the value 1 to gene A and also the value 1 to gene B because they are both involved in this closed walk. So to extract the real number of closed walks between a set a genes we have to divide the sum of the matrix's diagonale by the power its been raised to. Sum(closed walks gene A + closed walks gene B) = 2 --> 2/2=1 closed walk between those genes. This reasoning works for cycles only. A closed walk of length 2 or 3 is a cycle but a closed walk of length 4 isn't necessarily a cycle. Once the closed walks are extracted for each individual pathway and for the graph containing genes from both pathways we ordered the results in a dataframe. From the graph representing interactions in one pathway alone we extract the number of closed walk per gene. From the graph containing genes from both pathway we extract the total number of closed walks a gene perform, this number will not tell us how many of those closed walks are performed inside a pathway and how many are performed with genes from another pathway. This is why we extract the total number of closed walk to which we substract the number of closed walks performed in its own pathway. This gives us the number of closed walks performed between the 2 pathways so the number of cross talk! For each gene we have the number of closed walks it performs in its own pathway, and its number of crosstalks.
Results and discussion
Entry points: For two pathways, each gene performing crosstalk was plotted with the number of closed walks inside its own pathway he is involved in on the X axis and the number of crosstalks he is involved in on the Y axes. This allows us to highlight genes that are considered entry points, genes that are interacting much more than others. By looking at those "outliers" a little closer we realised that each of them was actually a transcription factor. PUT THEIR NAMES HERE. This also allows us to see between 2 pathways which one contains more communicating genes Meaning the number of ways back and forth is the same two pathways because one crosstalk belongs to both pathways. But for one pathway the crosstalks are involving less different genes than in the other (see big scheme).
Symmetry:
There is an asymmetry in term of communicating genes. Between 2 pathways performing together 100 crosstalks, the first can accomplish those with 10 genes only while the second has for example 40 genes performing those 100 crosstalk. But there is no asymmetry in the crosstalking process.
In order to know if two pathways need each other or if one is rather exploiting the other for stimuli we need to know which gene initiate a closed walk/cycle. We did not have this information. So with our analysis we can only know how many crosstalk happen between 2 pathways and so it is symmetric since a crosstalk is communication between 2 pathways. To know if one pathway needs another we should either look at who initiates the first interaction in each crosstalk or not look at crosstalks at all but simply compute the number of interactions going from one pathway to the other without looking for a return to its initiator. With this method we could see if one pathway gives or receives more stimuli from another.
The crosstalking process depends on the pair of pathways studied. We cannot conclude that a pathway crosstalks more than another. What we could determine is if a pair of pathways crosstalks more than another pair. Indeed we found that there is more crosstalk between cell death and cell differentiation than there is between cell death and mitotic cell cycle (only for cycles of length 2 and 3).
We only computed closed walks of length 2, 3 and 4. It is important to keep in mind that the labelling of the genes (in which pathway it is involved) is incomplete and biaised information. So talking in terms of pathways is not very precise. The separation of biological process into different pathways is a very human vision of things and we do not believe that it is representative of the reality. We can analyse closed walk as much as we want but if we don't know what gene/protein initiate the first interaction we can't know which pathway regulates the other. Suppressing the genes in the intersection of two pathways was necessary for the analysis but biologically irrelevant and leads to the loss of a lot of information. At the end of our work we concluded that genes should maybe not belong to a pathway but genes are more precisely included in cascade and maybe they should be labelled according to that. We think that if we broaden this analysis (higher length, more pathways, etc.) we would only conclude that everything is connected and can't be separated into pathways. This analysis from the point of view of each gene individually may be more accurate. For length 4 and more, the number of closed walks going back to a gene is dependant on how many genes are in the vincinity of that gene and how "well connected" it is to the rest of its pathway. In the end this is just describing that a pathway is more well-known than another pathway and that connections have been better established between the genes. This is a major bias in our analysis, as we take for reference an edgelist table that shows an asymmetry that might just exist because of insufficient knowledge.
Perspectives
Computing the number of interactions initiated by each gene and the number of interactions in which it is a target could allow us to build some sort of score where each gene is evaluated on its ability to regulate others and maybe built a map to find the highest "leaders".
We would like to thank our supervisor, Miljan Petrović, for this very stimulating project.