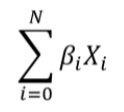Difference between revisions of "Polygenic Risk Scores"
Sbprm2021 3 (talk | contribs) |
Sbprm2021 3 (talk | contribs) |
||
| Line 15: | Line 15: | ||
=Jura & UK Biobank= | =Jura & UK Biobank= | ||
In order to have data, we used Jura, which is a cluster for analysis of sensitive data and is primarily used by the CHUV. We need to have authorization to use Jura, but like that we can have real subject’s data.And there are data coming from the UK Biobank, which is a large-scale biomedical database and research resource, containing genetic and health information from half a million UK participants that is enabling new scientific discoveries to be made that improve public health. | In order to have data, we used Jura, which is a cluster for analysis of sensitive data and is primarily used by the CHUV. We need to have authorization to use Jura, but like that we can have real subject’s data.And there are data coming from the UK Biobank, which is a large-scale biomedical database and research resource, containing genetic and health information from half a million UK participants that is enabling new scientific discoveries to be made that improve public health. | ||
| + | |||
| + | |||
| + | =Our goal with this project= | ||
| + | Our goal is to compare the polygenic risk score that we found for the SNPs of the different BMI indices from group 1. And if possible make connections and correlations with diseases. | ||
| + | |||
Revision as of 17:15, 4 June 2021
- Project name: Polygenetic Risk Score (PRS)
- Tutor: Alex Button (alexanderluke.button_AT_unil.ch)
- Slides: File:Polygenetic Risk Score.pdf
Contents
Introduction
Definition
Polygenic Risk Score or PRS is the calculation of a risk score from the subject's genome with the weighted sum of the SNPs to estimate the risk that the subject will develop a certain disease or trait according to the changes linked to the phenotype studied. There are two main factors that we use to calculate the PRS: the allelic dosage (Xi), which represents the number of copies of the main effect allele and can be 0, 1 or 2. And the other factor is the effect size (βi), which measures the strength of the relationship of a trait with the allelic dosage at a position i on a numeric scale.
Jura & UK Biobank
In order to have data, we used Jura, which is a cluster for analysis of sensitive data and is primarily used by the CHUV. We need to have authorization to use Jura, but like that we can have real subject’s data.And there are data coming from the UK Biobank, which is a large-scale biomedical database and research resource, containing genetic and health information from half a million UK participants that is enabling new scientific discoveries to be made that improve public health.
Our goal with this project
Our goal is to compare the polygenic risk score that we found for the SNPs of the different BMI indices from group 1. And if possible make connections and correlations with diseases.
Why is Polygenic Risk Score important ?
The Polygenic Risk Score is used for disease outcome prediction. It indicates the relative risk for the disease or trait studied, compared to other people with different genomes at an individual level. But we must be careful with the results, because it gives us correlations and correlations are not equal to causations. There are other factors that are not necessarily considered, such as the environment. Nevertheless it can be useful, for example: the riskiest people can be prepared and can take predispositions with their doctors. However, it will be more accurate for some people. For the moment, the use of PRS is not widespread, because there are no guidelines, and it is still in process of improvement. It will always be probabilities and not certitudes. Despite the statistics behind it, PRS could potentially be very powerful to “simply” predict (at birth) all kinds of diseases one could encounter. It is still a bit early for that, but it could help people in the future.