The domestic yak (Bos grunniens) is an important domesticated species for Tibetans. Domestic yaks provide meat and other basic resources of necessity. The analysis of yak genome provides important insights into adaptation to a high altitude. Here discussed study was published in Nature Genetics.
The study compares the yak genome with the genome of taurine cattle (B. taurus). Yak and cattle are cross-fertile, that means that they are genetically very similar. However the cattle suffer from hypertension when living in the yak habitat, thus, comparing this two species can provide the information about evolutionary adaptation to high altitude.
In the study, researches sequenced genome of a female yak. They found three genes that help the animal to deal with a low concentration of oxygen that is typical for high altitude. Five further genes provide a better nutritional assimilation, as a consequence of the limited herbal resources available in the mountains where they live.
 |
| Fig.1 Qiu et al., The yak genome and adaptation to life at high altitude., Nature Genetics 44, 2012 |
Venn diagram showing unique and shared gene families between the yak, cattle, dog and human genoms.
One the Fig.1 unique and shared gene families from four different species (yak, cattle, human and dog) are shown. Gene family is a set of genes that show sequence similarity, and generally (not always) have similar functions. We discussed the choice of the species on the Venn diagram. The two other species for comparison, beside of yak and cattle, were chosen probably because of the good annotated genome. It would be also nice to see the comparison to chimp, to see the relationships yak-cattle and human-chimp together. However the authors mention that yak and cattle have diverged approximately 4.9 million years ago, which is comparable to the time at which humans and chimpanzees diverged. Why the comparison is made to mammals and not other species? The comparison was done to show influence of adaptation, and for this purpose it is better to take more related species.
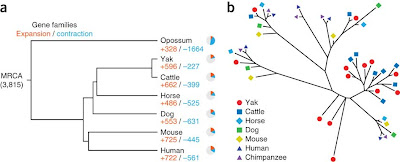 |
| Fig.2 Qiu et al., The yak genome and adaptation to life at high altitude., Nature Genetics 44, 2012 |
Gene expasion and contraction in the yak genome
In the Fig.2 a neighbor-joining tree of mammalian Hig domain sequences is presented. The Hig domain is known to play role in hypoxia, so adaptation to high altitude. Hig domain sequences group in different clusters, but yak and cattle sequences seem to be similar. Some brunches of the tree are long, that means that the sequences were very diverged.
Authors also show that yak has three more positively selected genes then cattle, and it is more then the rest of positive selected genes. The paper also compares Ka/Ks ratio of GO categories of yak and cattle. Ka/Ks ratio is a proportion of synonymous to nonsynonymous substitutions. It is assumed that synonymous substitutions represent the background and the selection influence nonsynonymous substitutions.
For the further studies it would be interesting to compare the results from other species, such as goats in mountains and fields, or even wild species. We wondered why there is now comparison of the results of the current study with already known result from the studies of human genome on adaptation to high altitude?
Summary
The study present analysis for the genome adapted to high altitude. The authors sequenced genome of yak using a whole-genome shotgun strategy and the Illumina platform. The scientist hope that this results can help in research of hypoxia-related diseases in humans.
Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, Auvil L, Capitanu B, Ma J, Lewin HA, Qian X, Lang Y, Zhou R, Wang L, Wang K, Xia J, Liao S, Pan S, Lu X, Hou H, Wang Y, Zang X, Yin Y, Ma H, Zhang J, Wang Z, Zhang Y, Zhang D, Yonezawa T, Hasegawa M, Zhong Y, Liu W, Zhang Y, Huang Z, Zhang S, Long R, Yang H, Wang J, Lenstra JA, Cooper DN, Wu Y, Wang J, Shi P, Wang J, & Liu J (2012). The yak genome and adaptation to life at high altitude. Nature genetics, 44 (8), 946-9 PMID: 22751099
