![]()
Phenotype convergence is a fascinating topic in evolution. Usually species evolve by divergence, starting from a common ancestor and then developing different genomic changes that lead to different phenotypes. which are then selected by the environment. Nevertheless, it has been observed in several examples that two or more different species, even very far-related in the phylogenetic tree, appear to have developed, after their divergence, similar phenotypic traits in order to adapt to the environment, therefore leading to an apparent convergence of their branches.
The aim of this work is to investigate the hypothesis according to which convergent phenotypes are not just a lucky coincidence produced by different point-mutations occurred in different species, but rather that a convergent phenotype is associated with the same mutation in all the species involved, and that these mutations are not happen by chance but are pushed by adaptation to the environment.
In order to do it, this group analysed sequence identities in the genomes of species that developed independently echolocation, certainly a very complex feature that it’s hard to believe it has developed in different species just by chance.
The first step was building the gene set to work on. Therefore, they sequenced the genome of four different bat species (both echolocating and non-echolocating), and acquired online the coding gene sequences (CDSs) of other bat species, a dolphin species and other non-echolocating animals (dog, cow, horse, mouse and human). In order to remove potential error sources, all ambiguous sites/codons were removed, and they selected only those genes that had no missing data and whose homologous were present in at least six of the investigated species. The final gene pool was about 2000 CDSs.
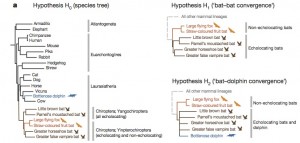
In order to evaluate the genetic convergence of these genes, they considered three different phylogenetic tree hypothesis.
H0: the real phylogenetic tree
H1: a fake phylogenetic tree in which all echolocating bats (in brown) are in the same branch
H2: a fake phylogenetic tree in which all echolocating animals (including dolphins) are in the same branch.
Then they proceeded with the alignment of all the CDSs in these species, and measured their SSLS
SSLS: site-wise log-likelihood support. A score that, based on the alignment of each amino acid in each gene of the investigated species, tells how much that alignment fits to a tree hypothesis.
If echolocating animals really have a sequence convergence in one or more genes, the alignment of these genes in echolocating animals will be better than expected, so it will fit to the fake phylogenetic trees (H1-H2) better than to the real tree.
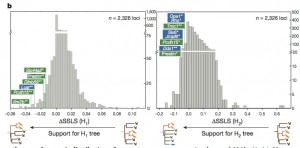
The fitness (SSLS) to the three trees is evaluated, and ?SSLS of each gene is calculated.
?SSLS: difference between SSLS to H0 and the SSLS to one of the fake trees. If the alignment of a gene fits more to H1 than to H0 (sequence convergence), then ?SSLS(H1) = SSLS(H0) – SSLS(H1) of that specific gene will be a negative number. This calculation is applied to all the genes of the pool. Then they checked the scores of those genes which were already found to be involved in echolocation.
All the hearing genes known to be involved in echolocation show negative ?SSLS, as well as other genes involved in hearing (green) and vision (blue), supporting the hypothesis of genetic convergence. With H2, the ?SSLS scores are still negative, but not so much, because the species involved are very distant and the hypothesis is more stringent.
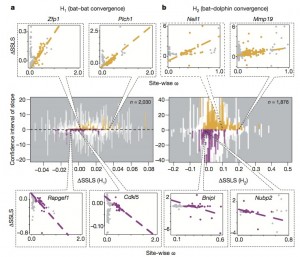
In order to evaluate whether or not these converging mutation were pushed by adaptation, they used the ? ratio as a score of the influence of selection for each site.
If ? > 1, it means that there is adaptation.
For each site, the ? score was correlated with ?SSLS. For instance, if in a gene ?SSLS and ? are inversely correlated (positive ? and negative ?SSLS) it means that adaptation has pushed those particular sites towards the same mutations in the different echolocating species.
As expectable, after screening all the gene pool, all possible kinds of correlations were found. What is interesting is that some genes involved in hearing and vision had a strong correlation between ? and ?SSLS that supports the hypothesis of convergence by adaptation (bottom left in the picture).
However, it’s not really specified if all the genes involved in echolocation showed this correlation.
The paper shows a very interesting approach for correlating genetic paths in evolution in different species, and some of the results strongly suggest the validity of the hypothesis of genetic convergence by adaptation. Nevertheless, it seems that the authors are trying to prove their point skipping some elements that would need to be further investigated.
First of all, it seems that negative ?SSLS is present also in many genes that haven’t shown, so far, any association with echolocation, a fact that might bring some doubts about how much the ?SSLS information is actually important in this context.
Moreover, in the second part of the paper, they show that one of the proteins with the best ? score is Cdk1, saying that it supports their hypothesis because this protein is important for the development of hair cells in the inner ear. But Cdk1 is a ubiquitary protein, necessary in the cell cycle of every type of cells, so it’s not a protein specifically involved in hearing.
Parker, J., Tsagkogeorga, G., Cotton, J., Liu, Y., Provero, P., Stupka, E., & Rossiter, S. (2013). Genome-wide signatures of convergent evolution in echolocating mammals Nature, 502 (7470), 228-231 DOI: 10.1038/nature12511