Meiosis is an important biological process by which combination of various types of the genes called alleles, are segregated and packed in each germ cell waiting to be transferred and expressed in descendants. This combinations of alleles are products of chromosomal crossovers (COs) during meiotic recombination, which increases the genetic diversity of gametes. Recombination may cause local mutagenic effect at crossover sites with recurrent double strand breaks (DSBs) and thus be the source of sequence variation too.
SUMMARY OF THE PAPER
By sequencing a large number of single sperm DNA molecules, the authors showed that meiosis is an important source of germline mutations and consequently gene variation. They found more de novo mutations in molecules with COs then in molecules without a recombination event by amplifying single CO products, using allele-specific PCR, at two previously identified recombination hotspots (HSI and HSII) from a pool of sperm. The binding site used by the human recombination machinery contains PRDM9 (PR Domain Containing 9), very polymorphic in humans. In order to investigate why sequence diversity positively correlates with high recombination activity regions, the authors sequenced 5,796 COs in total, including both reciprocal recombination products from 6 Caucasian donors. As a control they screened single nonrecombinants (NRs) in the same region and subset of donors using the same experimental conditions.
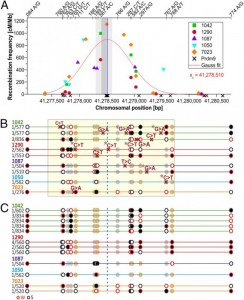
To adjust CO mutation frequencies, authors used mutation frequency of NRs as a control, which are combination of rare de novo mutations and PCR artefacts. COs had a mutation frequency nearly 3.6 times higher than NR control (0.29% de novo mutations per CO of which 50% occurred between the DSB and the CO breakpoint, 348 nucleotides away from hotspot center (Fig. 1A), and it was similar for both hotspots (HSI and HSII), but most of the donors actually came from HSI, so the authors focused more on data analysis of this hotspot.
Figure 1. COs, mutations, and CCOs in HSI.
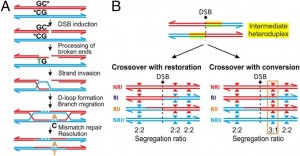
This also suggests that the observed mutations are associated with CO formation and they are independent of other site-specific factors such as base composition. Mutation rate at hotspots (? HS) showed that more active hotspots exert a stronger mutagenic effect than weaker hotspots. All the observed de novo mutations changed strong (S) CG into weak (W) TA base pairs and they all occurred mainly at CpG sites. This strong mutational bias at CpGs are not exclusive to COs. CpG dinucleotides generally have high mutation rates, but what explains high mutation level at COs containing CpG sites is that single stranded DNA arised from double strand break (DSB), is more susceptible for chemical modification leading to G ? T conversion on single-stranded DNA, and repair of those mismatched base pairs is only possible by the recognition from repair machinery bounded for double stranded, which is not the case for single stranded resected DNA 3’-ends created after DSB (Fig. 2A). If the formation of single-stranded DNA at methylated CpG sites is the main driver for de novo mutations, then DSBs resolved alternatively as noncrossovers (NCOs) might also have a higher frequency of the mutations.
Figure 2. Model of CO-driven evolution.
Considering hotspot CpG sites, the authors also observed very high methylation level (83-88%), both, in testis and in sperm, pointing on the cellular states before and after meiosis, respectively. The authors also suggested that non-Mendelian segregation of alleles at hotspots, arised during DSB repair could be either a result of an initiation bias, in which DSB-suppressing alleles are used to repair the broken homolog, or gene conversion favoring GC-alleles leading to GC-biased gene conversion (g-BGC).
Biased transmission is favorised for GC alleles representative for g-BGC, rather than for an initiation bias. In the authors data it is shown that all of the donors are homozygous at the DSB site which makes initiation bias unlikely. Sites with the strongest evidence for unequal transmission favor strong (GC) vs weak (AT) alleles (Fig 1B). Authors observed that transmission bias affects also single nucleotide polymorphisms (SNPs), which can be preferentially transferred, regardless of whether they are near or away from DSB (Fig.1C). Initiation bias do not favor strong over weak alleles in humans and yeast, whereas gBGC does. Another potential source of gBGC in the 6,085 mapped COs is the formation of COs with rare, discontinuous conversion tracks containing two CO breakpoints, and it is called complex COs (CCOs). Heteroduplex tracts are formed in polymorphic regions during DSB. If there acts gBGC, COs will tend to include strong alleles and exclude weak alleles (Fig. 2B). How fast crossing over drives the decimation of a hotspot via mutagenesis and gBGC depends on how sequence changes affect CO frequencies, which is still a mystery.
CONCLUSIONS
This study contributes to the understanding of the sequence evolution at recombination hotspots. Authors suggested that GC-biased gene conversion (gBGC) is the dominant force shaping the nucleotide composition at hotspots during crossing over, and potentially in other recombination products, which might explain the high GC content associated with recombination. It is possible that gBGC is an adaptation to reduce the mutational load of recombination, knowing that mutation favors weak over strong nucleotides. Still, small sample size gives a little power for detecting potential differences between COs and NCOs.
Arbeithuber, B., Betancourt, A., Ebner, T., & Tiemann-Boege, I. (2015). Crossovers are associated with mutation and biased gene conversion at recombination hotspots Proceedings of the National Academy of Sciences DOI: 10.1073/pnas.1416622112