Difference between revisions of "Modeling microbial ecosystem stability through metabolite leakage"
(Created page with "==Modeling microbial ecosystem stability through metabolite leakage== thumb ==Introduction== The plankton paradox, which highlights the unexpectedly high...") |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Modeling microbial ecosystem stability through metabolite leakage== | ==Modeling microbial ecosystem stability through metabolite leakage== | ||
| + | == Credits == | ||
| + | ''By Geiser Larissa and Besançon Léo'' | ||
| + | |||
| + | ''Supervisor: Snorre Sulheim'' | ||
| + | |||
| + | Click [https://github.com/GiveMe1Coffee/m6-consumer-resource-model here] to access the Github repository of the project. | ||
| + | |||
| + | Click [[Media:CRM_PPW_WIKI.pdf|here]] to download the powerpoint presentation | ||
| − | |||
==Introduction== | ==Introduction== | ||
The plankton paradox, which highlights the unexpectedly high diversity of plankton species despite limited nutrient sources, finds a parallel in the microbial world, particularly among bacteria with carbon sources. In theory, a finite number of carbon sources should lead to competitive exclusion, where only a few bacterial species dominate by outcompeting others for these resources. However, observations reveal a rich diversity of bacterial species coexisting, each seemingly thriving on similar or even identical carbon sources. This phenomenon suggests that, like plankton, bacteria benefit from various ecological mechanisms such as temporal fluctuations in resource availability, microhabitat diversity, and interactions like mutualism and predation. These factors contribute to a dynamic ecosystem where multiple bacterial species can coexist, utilizing the same carbon sources in ways that prevent competitive exclusion and support high biodiversity. | The plankton paradox, which highlights the unexpectedly high diversity of plankton species despite limited nutrient sources, finds a parallel in the microbial world, particularly among bacteria with carbon sources. In theory, a finite number of carbon sources should lead to competitive exclusion, where only a few bacterial species dominate by outcompeting others for these resources. However, observations reveal a rich diversity of bacterial species coexisting, each seemingly thriving on similar or even identical carbon sources. This phenomenon suggests that, like plankton, bacteria benefit from various ecological mechanisms such as temporal fluctuations in resource availability, microhabitat diversity, and interactions like mutualism and predation. These factors contribute to a dynamic ecosystem where multiple bacterial species can coexist, utilizing the same carbon sources in ways that prevent competitive exclusion and support high biodiversity. | ||
| Line 7: | Line 14: | ||
==Our project== | ==Our project== | ||
Our project consisted of modelling a Consumer-resource model of bacteria and carbon sources with a leakage parameter that can lead to byproduction of resources, which is one of the theory behind the plankton paradox. The model is based on the following differential equations: | Our project consisted of modelling a Consumer-resource model of bacteria and carbon sources with a leakage parameter that can lead to byproduction of resources, which is one of the theory behind the plankton paradox. The model is based on the following differential equations: | ||
| − | + | [[File:Equations.png]] | |
Then by exploring and analysing the model our goal was to determine in which situation and with which set of parameters there was a cohabitation between the species and no competitive exclusion. | Then by exploring and analysing the model our goal was to determine in which situation and with which set of parameters there was a cohabitation between the species and no competitive exclusion. | ||
Latest revision as of 12:26, 5 June 2024
Contents
Modeling microbial ecosystem stability through metabolite leakage
Credits
By Geiser Larissa and Besançon Léo
Supervisor: Snorre Sulheim
Click here to access the Github repository of the project.
Click here to download the powerpoint presentation
Introduction
The plankton paradox, which highlights the unexpectedly high diversity of plankton species despite limited nutrient sources, finds a parallel in the microbial world, particularly among bacteria with carbon sources. In theory, a finite number of carbon sources should lead to competitive exclusion, where only a few bacterial species dominate by outcompeting others for these resources. However, observations reveal a rich diversity of bacterial species coexisting, each seemingly thriving on similar or even identical carbon sources. This phenomenon suggests that, like plankton, bacteria benefit from various ecological mechanisms such as temporal fluctuations in resource availability, microhabitat diversity, and interactions like mutualism and predation. These factors contribute to a dynamic ecosystem where multiple bacterial species can coexist, utilizing the same carbon sources in ways that prevent competitive exclusion and support high biodiversity.
Our project
Our project consisted of modelling a Consumer-resource model of bacteria and carbon sources with a leakage parameter that can lead to byproduction of resources, which is one of the theory behind the plankton paradox. The model is based on the following differential equations:
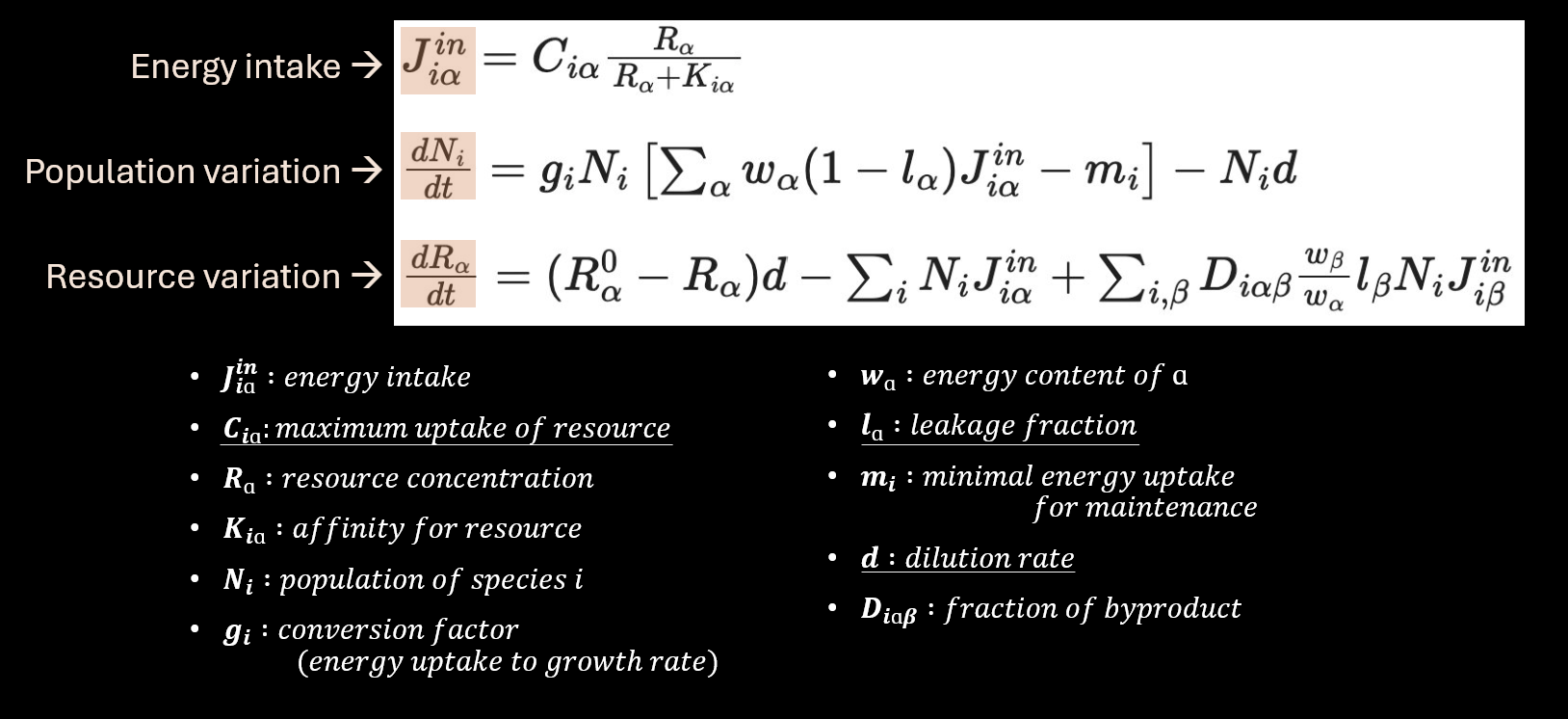
Then by exploring and analysing the model our goal was to determine in which situation and with which set of parameters there was a cohabitation between the species and no competitive exclusion.
the second goal was the determine what happens when we remove a specie.
Methods and Results
Our First approach of the problem after having created the model basically making variation manually in the parameters and try to understand how the model responded to it. Most of the parameters were initially set to some basic number like 0 or 1, we started to play with 4 different species and 10 different carbone sources. But we rapidly realized that it was way to much parameters to hope finding something manually so to try and grasp a better understanding of the dynamics of the model we used a simplified model by keeping the most important parameters as Larissa presented you before. We lowered the number of species to 3 and the number of carbone sources to 3 also and played manually with the C matrix and the D matrix, so with the maximum uptake of a resource for a species and the byproduction. We noticed that in some cases there was only one of the 3 species survived and on some other 2 species cohabitated. This led us to our second approach.
Our observations led us to pose the hypothesis that the cohabitation was possible when the conversion factor g times the growth intake is equal between the species that cohabit. In order to test if this hypothesis was true in all cases we decided to use the approach of randomness with statistical tests. Basically we made a large number of simulations where we randomly variated g, C, and D and then we saved the parameters of the simulations where there was cohabitation in a dataframe and we also saved the parameters of the simulations where there was a domination on one species in a different dataframe. We did 1000 simulations to ensure a good statistical power. Then we proceed to calculate for the species the growth intake and the multiplication of the g parameters with it. In order to determine if there was some statistical differences between the simulations we ran some Wilcoxon paired tests between different combinations of simulations. We used Wilcoxon paired test because the distribution of the G*growth_intake parameter was not normally distributed and the paired is because we need to keep in mind that each simulations has a complete different set of parameters and that’s why the data needs to be considered paired for the species that are in the same dataframe.
We tested 4 specifics situations. In cohabitation cases, Species cohabitating between each other. In domination cases, species dead against each other. In cohabitation cases, species alive vs species dead And finally in dominations cases, specie alive versus species dead.
The results of the test were the following. In both cases of Alive vs Alive and Dead vs Dead the pvalue was not significant which suggest that the there is no real difference between the species at these states. But when we compared the species Alive against the dead ones we obtained significant p-values in the cohabitation dataset as well as in the domination dataset, suggesting this time a clear difference in the combination of parameters between Alive and dead species. These results infere that our hypothesis is correct.
These was expected because if there is a cohabitation it means that the species are at the equilibrium and that therefor their dN/dt = 0 and that g*growth_intake = d (dilution rate).
Our 3rd approach was to simplify the model with only 2 species, one carbon source at first and a 2second generated by byproduction. We calculated the theoretical values For R1, R2, N1 and N2 at the equilibrium state mathematicaly with the equations.
then for a controlled range of values we variated the leakage of the first species and the dilution rate, then for every combination of the two we calculated the theoretical density of the ressources R1 and R2 at equilibrium and made the simulation with our model. Then we computed the difference between the results of the simulations and the expected theoretical results. (see the powerpoint slides) The Goal of doing that is to find where, if so, there is a difference between the simulations and the theory, which could mean that the simulation model collage at these specific numbers. Our goal was to look for threshold in the parameters where the model collapse from the theory. After analysing these tresholds for the ressources we can at the evolution of the species density within these tresholds.
The Results allowed us to say that the leakage appeared to be an essential parameter for the coexistance of species with limited ressources.
Perspectives
We unfortunatly ran low on time and couldn't go further with our analysis, the next step would have been to expend the model with a second cross-feeding resources coming from the second species and redo the math, then trying to apply the tresholds to the complete model.