ExpressionView
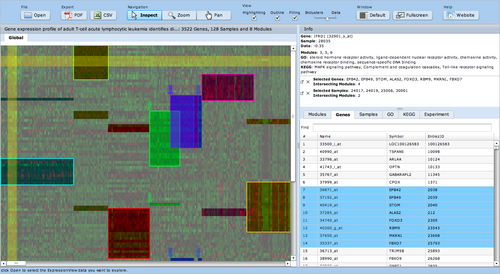
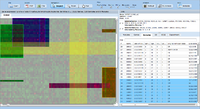
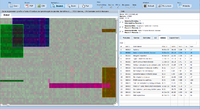
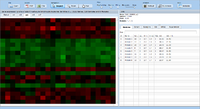
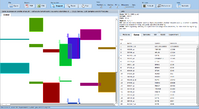
ExpressionView is an R package that provides an interactive environment to explore biclusters identified in gene expression data. A sophisticated ordering algorithm is used to present the biclusters in a visually appealing layout. From this overview, the user can select individual biclusters and access all the biologically relevant data associated with it. The package is aimed to facilitate the collaboration between bioinformaticians and life scientists who are not familiar with the R language.
Launch and download
Launch the ExpressionView Flash applet
- Launch ExpressionView
- Launch ExpressionView with adult T-cell acute lymphocytic leukemia (ALL) data (8 modules)
- Launch ExpressionView with adult T-cell acute lymphocytic leukemia (ALL) data (108 modules)
Download the R package (includes the Flash applet)
- Windows binary package
- Mac OS X 32 bit binary package, Mac OS X 64 bit binary package
- Source package
Download the stand-alone viewer (adobe AIR)
If you prefer a stand-alone viewer, you can download and install the Adobe AIR build ExpressionView.air (right-click to download file).
To run the program, you need the AIR runtime environment which you can get from Adobe.
ExpressionView creates files associations to .evf files, allowing you to simply double-click on such files to launch the viewer and load the data.
Download sample data
- Gene expression profile of adult T-cell acute lymphocytic leukemia (ALL) with 8 modules (right-click to download file).
- Gene expression profile of adult T-cell acute lymphocytic leukemia (ALL) with 108 modules (right-click to download file).
Screenshots
Tutorials
There are several tutorials describing how to use ExpressionView:
R package
Flash applet
Quick help (pdf) Getting started (video tutorial) Using the tables (video tutorial) Modular view (video tutorial) View and Export functions (video tutorial)