![]() How do adaptive phenotypes evolve? This question, despite the increasing availability of genomic and other molecular data, remains still largely unanswered. Among the different aspects investigated, a major point of discussion in this topic is the extent of the contribution of coding versus non-coding variation in the evolution of new traits. Although many research groups suggested that non-coding mutations might play a pivotal role because might avoid pleiotropic effects, still few examples are available to discard a potential major contribution of coding variants in adaptive evolution.
How do adaptive phenotypes evolve? This question, despite the increasing availability of genomic and other molecular data, remains still largely unanswered. Among the different aspects investigated, a major point of discussion in this topic is the extent of the contribution of coding versus non-coding variation in the evolution of new traits. Although many research groups suggested that non-coding mutations might play a pivotal role because might avoid pleiotropic effects, still few examples are available to discard a potential major contribution of coding variants in adaptive evolution.
The paper from Jones et al. we discussed tried to answer this question by looking at the differences between distinct populations of threespine sticklebacks (Gasterosteus aculeatus). This species, originally found in marine habitats, colonized the freshwater environment evolving specific phenotypic traits, but still maintaining the ability to hybridize with the marine individuals. An important feature of this species, already known from previous studies, is the presence of shared genomic variants in geographically unrelated populations distinguishing the marine from the freshwater populations. This finding suggested the possibility of a parallel adaptive evolution of phenotypic traits due to the reuse of standing genetic variation. To test this hypothesis, Jones et al. generated a reference genomic assembly of a female freshwater stickleback (Sanger sequencing, 9.0x coverage, total gapped size: 463Mb). This reference genome provided the basis to analyze genomic differences in marine and freshwater populations collected in several locations around the world (Europe, North America and Japan). For this purpose, a total number of 20 individuals (classified in clearly marine and clearly freshwater based on multiple phenotypic features) were sequenced at 2.3x average coverage and genome-wide single nucleotide polymorphisms (SNPs) identified.
The data collected were analyzed using three different approaches with the aim of finding regions in the genomes showing a high similarity among the freshwater individuals and differing from the corresponding loci in the marine samples. The first approach consisted in a self-organizing map-based iterative Hidden Markov Model (SOM/HMM), used to reconstruct common relationships (trees) among the individuals. Although most of the phylogenies recapitulated the geographical relationships among the samples, four of them separated most of the marine from most of the freshwater individuals, identifying genomic loci putatively involved in the differentiation of the ecotypes. The second and the third approaches used a sliding window analysis to detect the divergence between the two populations. The second consisted in the calculation of a cluster separation score (CSS) to quantify the distance between the marine and the freshwater clusters; the third consisted in an unguided Bayesian model-based data-driven clustering (DDC) to calculate for each window a maximum number of clusters to which assign the individual samples. In total, 242 genomic regions showing a shared marine-freshwater divergence were identified by either method (0.5% of the genome). Testing of these approaches on a genomic location known to have evolved adaptively in the distinct species (EDA gene, fig.1) revealed the reliability of the three and the power of their complementary usage to spot putatively adaptively evolving loci.
Figure 1: Parallel divergence signals at known armour plate locus. a) Ensembl gene models around EDA. b) Visual genotypes for sequenced fish (homozygous sites for most frequent allele in marine fish (red); homozygous for alternative allele (blue); heterozygous (yellow), or non-variable/missing/repeat- masked data (white)). c) DDC cluster assignments for marine (red) and freshwater populations (blue). Most fish are assigned to cluster k1, except in the boxed region, where freshwater fish are assigned to a distinct cluster (k2). d) SOM/HMM analysis supports patterns of divergence with a marine– freshwater-like tree topology in the centre, but not edges, of the window (trees a–d). e, f) Similar support is shown by CSS analysis (e) and its associated P-value (f). The combined analyses define a consensus 16-kb region shared in freshwater fish (vertical shaded box), matching the minimal haplotype known to control repeated low armour evolution in sticklebacks.
To test the extent of parallel reuse of these regions in adaptation to the freshwater environment in contrast to newly evolved adaptive loci, an independent sample of a pair of marine and freshwater individuals from the same geographical zone (River Tyne, Scotland) was subjected to sequencing and SNP analysis. The experiment showed that, within the most highly divergent windows of the genome, only a part (35.3% of the 0.1% most divergent windows) contained the globally shared loci (fig. 2). The result indicates that part of the divergence between the two phenotypes actually derives from shared standing variation, but also that new population specific mutation can play a role in the determination of the specific traits.
Figure 2: How much of local marine–freshwater adaptation occurs by reuse of global variants? a) Classic marine and freshwater ecotypes are maintained in downstream and upstream locations of the River Tyne, Scotland, despite extensive hybridization at intermediate sites16. b) Pairwise sequence comparisons identify many genomic regions that show high divergence between upstream and downstream fish (x axis). Many, but not all, of these regions also show high global marine–freshwater divergence (y axis; red points indicate significant CSS FDR , 0.05), indicating that both global and local variants contribute to formation and reproductive isolation of a marine– freshwater species pair.
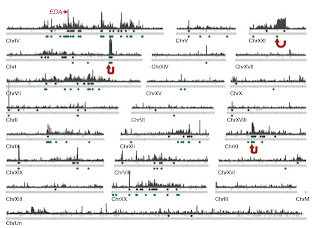
Interestingly, the group found three loci showing clear marine-freshwater divergence within regions involved in chromosomal inversions (chromosomes I, XI and XXI, fig. 3). The finding supported the hypothesis that molecular mechanisms, such as chromosomal inversions, suppressing recombination between adaptive loci can be favored by selection for the maintenance of contrasting ecotypes in hybridizing populations.
Figure 3: Genome-wide distribution of marine–freshwater divergence regions. Whole-genome profiles of SOM/HMM and CSS analyses reveal many loci distributed on multiple chromosomes (plus unlinked scaffolds, here grouped as ‘ChrUn’). Extended regions of marine–freshwater divergence on chromosomes I, XI and XXI correspond to inversions (red arrows). Marine–freshwater divergent regions detected by CSS are shown as grey peaks with grey points above chromosomes indicating regions of significant marine– freshwater divergence (FDR , 0.05). Genomic regions with marine– freshwater-like tree topologies detected by SOM/HMM are shown as green points below chromosomes.
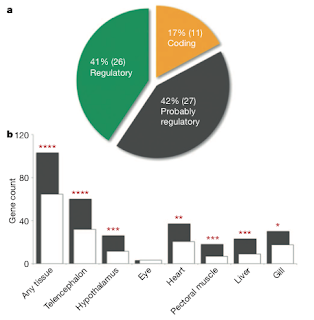
Finally, the analysis of the 64 genomic regions showing the strongest evidence of parallel evolution were investigated to determine the contribution of coding and non-coding variation to the adaptation to a different environment. Only 17% of them could be classified as coding based on the presence of non-synonymous substitutions, while the remaining part could be attributed to regulatory or probably regulatory changes (fig. 4a). To actually test if any regulatory change could be linked to these regions, a whole-genome microarray expression analysis was performed on tissues from a marine and a freshwater sample. The results obtained from genes mapping within or close to the loci identified by either method show a general divergence in the expression levels in different tissues between the two ecotypes (fig. 4b).
Figure 4: Contributions of coding and regulatory changes to parallel marine–freshwater stickleback adaptation. a) A genome-wide set of marine– freshwater divergent loci recovered by both SOM/HMM and CSS analyses includes regions with consistent amino acid substitutions between marine and freshwater ecotypes (yellow sector); regions with no predicted coding sequence (green sector); and regions with both coding and non-coding sequences, but no consistent marine–freshwater amino acid substitutions (grey). b) Genome-wide expression analysis shows that marine–freshwater regions identified by SOM/ HMM or CSS analyses are enriched for genes showing significant expression differences in 6 out of 7 tissues between marine LITC and freshwater FTC fish (observed, grey bars; expected, white bars; *P , 0.01, **P , 0.001, ***P , 0.0001, ****P = 0.00001), consistent with a role for regulatory changes in marine–freshwater evolution.
In summary, the work provided strong evidence that in threespine sticklebacks many loci implicated in adaptation from marine to freshwater environment were reused independently in several distinct populations, suggesting parallel evolution had a deep impact in the adaptation process. Furthermore, several putatively adaptive loci were found to be involved in chromosomal inversions, supporting the idea that genomic rearrangements can hamper recombination in these genomic locations thus preserving the features of the different ecotypes. Finally, the results obtained suggest regulatory mutation might have had a major role in the evolutionary processes leading to the adaptation of this species to a new environment.
Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, Birney E, Searle S, Schmutz J, Grimwood J, Dickson MC, Myers RM, Miller CT, Summers BR, Knecht AK, Brady SD, Zhang H, Pollen AA, Howes T, Amemiya C, Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team, Baldwin J, Bloom T, Jaffe DB, Nicol R, Wilkinson J, Lander ES, Di Palma F, Lindblad-Toh K, & Kingsley DM (2012). The genomic basis of adaptive evolution in threespine sticklebacks. Nature, 484 (7392), 55-61 PMID: 22481358