A biochemical story on convergent evolution
Introduction
Convergent evolution is the process by which similar traits evolve independently in distantly related organisms, such as wings in bats and birds. This can target orthologous or unrelated genes, which gives a different view on the concept of convergent evolution : how much it is constrained to some pathways, or, reversely, how diverse the path to the same function can be.
For convergent evolution to arise, different proteins must be assembled into an ordered, functional pathway. Currently, Three hypotheses shed light on the matter. Under the cumulative hypothesis, enzymes catalyzing the earlier reactions of a pathway must evolved first, because, otherwise, enzymes that perform the following steps would have no substrate to react with. Later steps would arise by duplication of the first enzyme. This suppose that intermediates are advantageous. Reversely, under the retrograde hypothesis, enzymes catalazing the later steps of a pathway would evolved first, and then gene duplication would give rise to the enzymes catalysing earlier steps. This suppose that intermediates could be produced non-enzymatically but doesn’t assume anything on their potential effect. Finally, the patchwork hypothesis states that a novel pathway will arise by the recruitment and rerouting of an alternative, preexisting pathway – we talk about ‘exapted’, or ‘co-opted’ enzymes. This suppose that the older, recruited enzyme was catalazing a promiscous reaction.
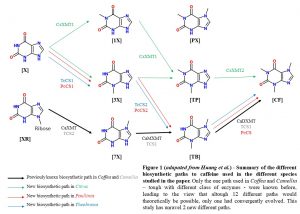
In plants, one of the most studied example of convergent evolution is caffein biosynthesis, which seems to have independently appeared at least five times during flowering plant history : only a few representatives of each clade display caffein biosynthesis, wich means, under the parcimony rule, that rather to be a trait ancestrally shared, it is more likely to have independently and repeatedely emerged. For the past 30 years, only one biosynthetic path among the several possible was shown to have convergently eveloved (Fig. 1), though with paralougous enzymes in both Coffea (XMT) and Camillia (CS) from the SABATH family. Most of those enzymes actually catalyze O-methylation, but were recently co-opted in those species to catalyze N-methylation.
In their article, Huang et al. [1] shed light in how caffeine convergence occured in 5 different species : Theobroma, Paullinia, Citrus, Coffea and Camillia. They were able to uncover different biosynthetic paths, thereby contradicting the idea that convergence in caffeine biosynthesis was constrained to one and only path, and to reconstruct ancestral enzyme activities, thereby illustrating to a molecular level how a new function can arise.
Different biosynthetic pathways to caffeine exist in Theobroma, Paullinia, and Citrus
To uncover genes and pathways used in plants to produce caffeine, Huang et al. first identify SABATH enzymes from each species studied and mapped them to the EST database to uncover the ones expressed in the caffeine producing tissues. Next they conducted enzyme assays to identify substracts with which they were able to react and mass spectroscopy scans to identify products, reconstructing full pathways to caffeine in those distantly related species (Fig. 2A).
Indeed, they could reveal that Theobroma and Paullinia express CS-type enzymes orthologous to the ones expressed in Camellia in their caffeine producing tissues. Surprisingly, they catalyze a different biosynthetic path (Fig. 1). Importantly, the enzymes catalyzing the first steps, TcCS1 and PcCS1 for Theobroma and Paullinia respectively, and the second step, TcCS2 and PcCS2 are respectively more distantly related than are TcCS1 and TcCS2, and PcCS1 and 2, which was unexpected considering their catalytic similarity. Therefore, it represents a strong evidence towards convergent repeated duplication of those enzymes in each lineage, rather than ancestrally duplicated enzymes that would have been lost in other non-caffeine producing lineages. However, tough Huang et al. provide a phylogenetic tree of SABATH enzymes with bootstrap values for major nodes, the ones providing that statement are not supported, which would have give further credit to it.
Because of the phylogenetic proximity of Paullinia and Citrus, that are both part of the Sapindales family, one could expect that they share the same enzyme type involved in caffeine production. Nevertheless, Citrus do not express CS-type enzymes in sites of caffein prduction but rather express two recently duplicated XMT-type enzymes ortologous to the ones found in Coffea, but they are specialized in another biosynthetic path to caffeine (Fig. 1).
Therefore, contrary to what has been believed for more than 30 years, plants have a much broader biosynthetic repertoire than previously known, with at least three different paths leading to caffeine biosynthesis that convergently emerged. However, this is unclear which proteins were exapted and what function they previously served, allowing them to be preserved along million of years of evolution.
Ancestral XMT enzymes displayed O-methylation
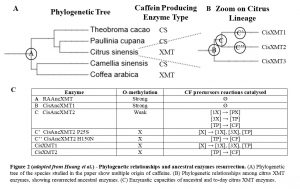
Coffea and Citrus XMT enzymes ancestors needed to be maintained for more than 100 My from their common ancestor, to then independently give rise to N-methylating enzymes involved in caffeine production. To understand what allow them to be maintained, Huang et al. used a method allowing to ‘resurect’ ancient protein [2]. This consist in inferring ancestral sequences based on to-day descendant protein alignments and to synthetize them to characterize their function.
Using that method, Huang et al. ressurect the 100 My old XMT enzyme ancestor to Rosids and Asterids, hereby called RAAncXMT (Fig. 2A), and its descendant CisAncXMT1, at the node giving rise to the citrus lineage (Fig. 2B). They both exhibit high O-methylation activty (Fig. 2C), which explains why they would have been maintained over such a long time, but no N-methylation activity. It is still the case of one of its to-day descendants in Mangifera, whereas they have specialized in N-methylation in Citrus. Today, Citrus possess a SAMT enzyme capable of both methylations, which could account for the loss of that function in XMT enzymes.
They also resurrected CisAncXMT2, at the node giving rise to both to-day CisXMT1 and 2, responsible for caffeine production. Interrestingly, CisAncXMT2, while still maintaining small O-methylation activity, display N-methylation activity, including almost all of the activities of both to-day enzymes, reconstituting together the two last steps needed for caffeine production, tough it is still unclear how it was recruited to form a functional pathway.
Ancestral citrus XMT enzymes were only a few steps away from to-day caffeine production function
To understand how nowadays Citrus XMT enzymes arised from CisAncXMT2, Huang et al. mapped it against CisXMT1 and 2 and identified key mutations. They then mutagenized the resurrected enzyme. In the lineage leading to CisXMT2, they identified one key mutation, P25S, that was sufficient to reproduce qualitatively the activity of CisXMT2 (Fig. 2C). Similarly, in the lineage leading to CisXMT2, they identified H150N as the mutation sufficient to reproduce roughly today’s activity. Altough other mutations could have shifted the ancestral enzyme activity, this shows that, after duplication, from those 2 single mutations alone, a complete pathway to caffeine would have emerge.
Two very interesting points sould be noted here. First, that very few mutations are sufficient to shift one enzyme substrate preference, which may have been a more widespread fact during evolution. Second, that contrary to the very linear biosynthetic vision one may have, several activities can emerge at the same time, and, more importantly, while maintaining the original activity, as it was the case for CisAncXMT1, thereby reconciling several hypothesis.
Conclusion
Using the very concrete exemple of caffeine Biosynthesis, Huang et al. were able to nicely illustrate the mechanisms of convergent evolution, unveiling much more diversity than previously thougth in the biosynthetic path, and to give us a view on the transition from the ancestral enzyme to the nowaday ones, demonstrating that the hypotheses running in the field were not mutually exculsive since biological pathways are not as linear as one may think.
Altough enzymatic data were quite strong and the whole story quite convincing, the phylogenetic analysis leading to the resurrection of enzymes would have benefit from the authors sharing statistical confidence on the alignment, and especially in the sites they mutagenised thereafter. Nevertheless, the study Huang et al. conducted was well constructed and easily understandable form people outside of the field, and we hope to learn more about the other caffeine producing plants, such as Guayusa, that contains much more caffein that coffea itself.
[1] Huang R, O’Donnell AJ, Barboline JJ, Barkman TJ (2016) Convergent evolution of caffeine in plants by co-option of exapted ancestral enzymes. Proc Natl Acad Sci USA 113:10613–10618
[2] Thornton JW (2004) Resurrecting ancient genes: Experimental analysis of extinct molecules. Nat Rev Genet 5(5):366–375