How exactly do lineages diverge to the point that they can be considered separate species, and especially reach reproductive isolation, is still an ongoing question in evolutionary biology.
Classical views of speciation hypothesize the existence of speciation genes, defined as loci contributing to reproductive isolation (Nosil and Schluter 2011). Although such loci have been clearly described in a few cases (Presgraves 2010), the mechanisms remain impossible to generalize. Two central issues remain unresolved:
- Whether genetic incompatibilities arise mostly through adaptive or neutral mutations;
- Whether these isolating mutations occur at specific loci in the genome.
In a recent paper, Hans Ellegren and his colleagues at Uppsala University describe the new insights brought by the comparison of whole genome data from two sister species of Old World flycatchers. The collared flycatcher Ficedula albicollis and the pied flycatcher F. hypoleuca (fig. 1) have been separate species for about 2 millions years, and their reproductive isolation is still incomplete as they can produce fertile male hybrids (but infertile females). Consistently, gene flow is strongly reduced but not null between the two species.
Photographs: Johan Träff. Nature 491, 756–760.
The researchers first established a map of the flycatcher genome by assembling short reads from a collared flycatcher and using the known zebra-finch genome as a reference to order and orient the flycatcher sequences. This flycatcher map was then used as a reference to reconstitute the genomic sequences of 10 unrelated males in each species.
The researchers then compared these 20 sequences to identify sites where the DNA sequence varied between at least two individuals, the so-called “single nucleotide polymorphisms” or SNPs. This comparison showed that a large proportion of the genomic variation was shared between the two species. Indeed, 53.4% of the variable sites in pied flycatchers and 38.6% in collared flycatchers were shared with the other species: both species varied at these sites.
To determine the basis for species divergence, the team focused on sites that varied between species but were identical for all the individuals of the same species. It turned out that these “sites of fixed divergences” were distributed non-randomly in the genome. In almost each chromosome, one or a few regions exhibited a proportion of fixed differences up to 50 times higher than the genome-wide mean (e.g. chromosome 1A in fig. 2). Such divergence peaks were mostly located at the ends or in the middle of chromosomes (telomeres and centromeres), both regions involved in cell division. Therefore, the separation of the species might be mainly based on changes in chromosome structure that would affect the production of germ cells, rather than on isolated adaptations of proteins.
Moreover, almost all these divergence peaks showed reduced diversity within both species, a signal that directional selection acted in the same regions in the two lineages. However, selection seemed to act on non-coding regulatory regions rather than directly on protein-coding genes: protein-coding genes in divergence peaks were more likely to show differences in expression between both species, but did not exhibit a stronger signal of selection in their DNA sequence.
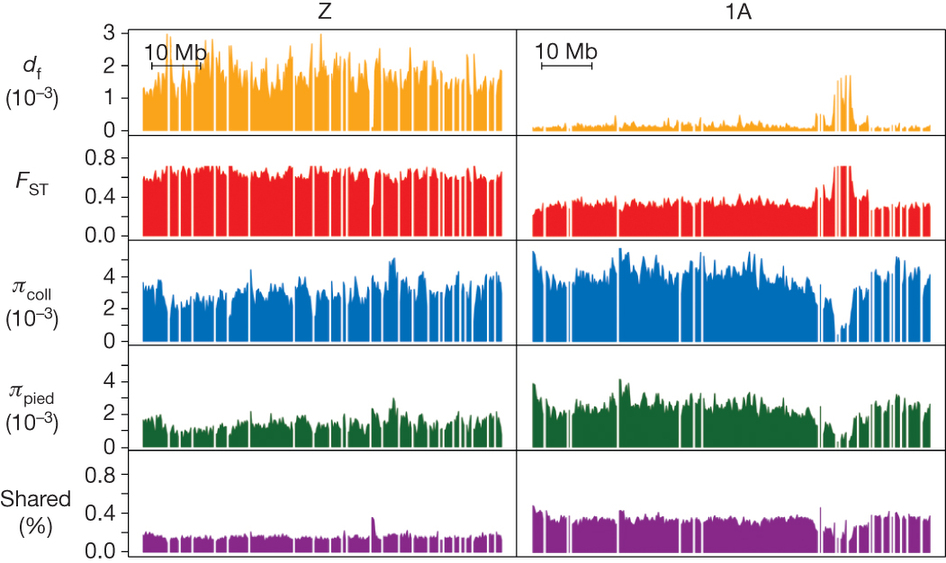
Interestingly, the pattern of differentiation was quite different on the Z chromosome, one of the sexual chromosomes in birds: males carry two copies, whereas females carry one copy and a W chromosome. The proportion of fixed differences was quite uniform all over the Z chromosome and amounted to that found in divergence peaks on autosomes (fig. 2). Divergence was thus much higher: overall, the Z chromosome contained about 35% of all fixed differences in the male genome. Hypothetically, that pattern could represent a second stage of species differentiation, where an increased background level of divergence would have blurred the divergence peaks. Whether such a pattern would appear through an accumulation of fixed differences between peaks or through an increase in peaks’ lengths remains an open question.
Taken together, these results suggest that reproductive isolation occur mainly through the accumulation of between-species differences in regulatory regions and/or regions involved in cell division. They confirm a key role for sex chromosomes in separating two lineages. The fact that no particular gene function seems to be strongly under selection hints at a first divergence step through neutral mutations, then followed by selection for differences in structural sites that avoid the production of hybrids with lower fitness. Studies of other pairs of sister species should now tell whether these conclusions could be generalized.
Ellegren H, Smeds L, Burri R, Olason PI, Backström N, Kawakami T, Künstner A, Mäkinen H, Nadachowska-Brzyska K, Qvarnström A, Uebbing S, & Wolf JB (2012). The genomic landscape of species divergence in Ficedula flycatchers. Nature, 491 (7426), 756-60 PMID: 23103876
