Development and homeostasis of all organisms is tightly controlled by transcription regulatory factors that are often highly conserved across deep phylogenies. However, it is unclear to what extend the basic components of these networks (e.g. network motifs and structure, binding frequencies, factor interactions) are preserved in distantly related species. Boyle and colleagues try to shed light on this question in a recent study published by nature in August 2014 (doi:10.1038/nature13668).
Paper summary
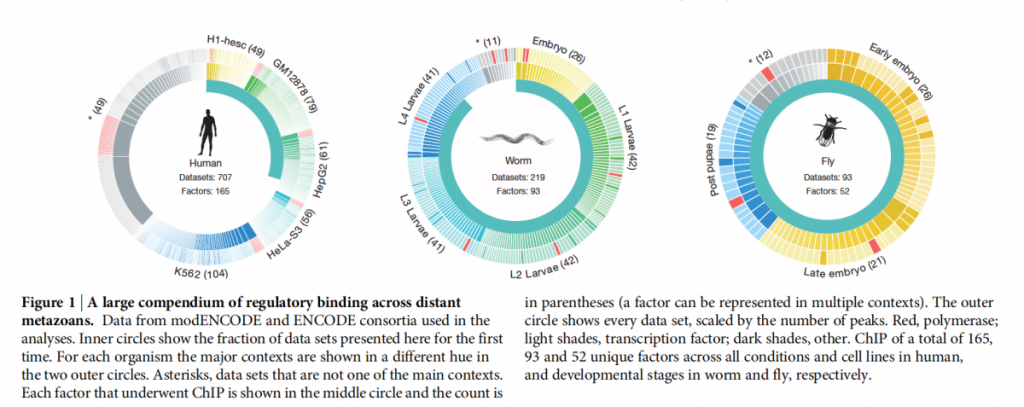
The scientists compare genome-wide binding locations of 165 human, 93 worm and 52 fly transcription regulatory factors in different cellular contexts (developmental stages and tissues) to identify the common properties of their underlying networks (data overview shown in Figure 1, taken from original publication).
As already described in smaller-scale studies, they see that DNA binding motifs of orthologous regulatory factors remain similar in distantly related species. Furthermore, these orthologous factors are expressed in similar contexts. However, expression of the orthologous targets is only weakly correlated suggesting an extensive re-wiring of regulatory networks across human, worm and fly. Reconstructions of regulatory networks point to a higher number of master-regulators and upward-flowing edges in human when compared to worm and fly. In all three species, the most abundant network motif is the feed-forward loop while cascade, divergent and convergent network motifs are underrepresented. Investigation of co-associations of regulatory factors shows that co-associations are local and contextual and that co-associations occurring at promoters have stronger conservation than co-associations in more distal regions.
The authors conclude that the overall structure of regulatory networks, in terms of network motif usage and context of the genomic binding event (high-occupancy target regions, enhancer, promoter), is strongly conserved. However, the regulatory targets are quite divergent and may account for the phenotypic differences among species.
Personal comment
Boyle and colleagues have created a big data set of transcription factor binding events in different cell lines and tissues for human, worm and fly. Doubtless, this resource will be of high interest for many researchers. But in my opinion, there are a couple of points that the reader should keep in mind when interpreting this data in light of the papers’ research aim – the comparison of regulatory circuits across species:
1) Tissue sampling
Without any question, cell lines are a good tool to study basic principles of cell homeostasis. But how well does a cell line, which has undergone many cell divisions and freezing/thawing cycles in an artificial lab environment, represent its in vivo counterpart? Is it reasonable to compare highly specialized human cell lines to entire worm and fly embryos? Why not use mouse instead of human? From an evolutionary perspective, mice are as close to worms or flies as humans are, and samples from an in vivo organ would probably reflect “reality” much better…
2) Genome size and complexity
The authors detect a higher number of master-regulators and upward edges in human regulatory networks when comparing human to worm and fly. But again, we are comparing highly specialized cell lines to whole embryos and larvae. Any signal created by specific tissues in an embryo or a larva has probably been strongly diluted and as a consequence, may be difficult to pick up! And not to forget: Human, worm and fly have different genome sizes and gene numbers. Anything we compare should be scaled and discussed in light of these numbers.
Big science can be helpful, but interpretation needs to be done carefully. Data is often generated based on very broad and general research questions and the generated data may not necessarily be the best to answer these questions. I am not against big consortia such as ENCODE and modENCODE, but I think that many of the findings can only be discussed from a very general perspective and often require a more redefined analysis.
Reference
Boyle, A., Araya, C., Brdlik, C., Cayting, P., Cheng, C., Cheng, Y., Gardner, K., Hillier, L., Janette, J., Jiang, L., Kasper, D., Kawli, T., Kheradpour, P., Kundaje, A., Li, J., Ma, L., Niu, W., Rehm, E., Rozowsky, J., Slattery, M., Spokony, R., Terrell, R., Vafeados, D., Wang, D., Weisdepp, P., Wu, Y., Xie, D., Yan, K., Feingold, E., Good, P., Pazin, M., Huang, H., Bickel, P., Brenner, S., Reinke, V., Waterston, R., Gerstein, M., White, K., Kellis, M., & Snyder, M. (2014). Comparative analysis of regulatory information and circuits across distant species Nature, 512 (7515), 453-456 DOI: 10.1038/nature13668