Introduction
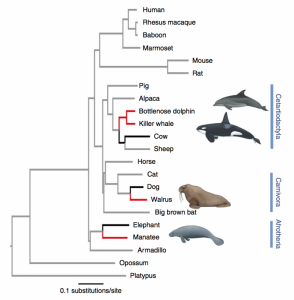
Convergent evolution is the independent evolution of similar features in species of different lineages. Marine mammals from different mammalian orders share several phenotypic traits adapted to the aquatic environment is a very classic example of convergent evolution. Although there are potentially several genomic routes to reach the same phenotypic outcome, it has been suggested that the genomic changes underlying convergent evolution may to some extent be reproducible and that convergent phenotypic traits may commonly arise from the same genetic changes. To investigate convergent evolution at the genomic level, the authors present high-coverage whole-genome sequences for four marine mammal species: the walrus (Odobenus rosmarus), the bottlenose dolphin (Tursiops truncatus), the killer whale (Orcinus orca) and the West Indian manatee (Trichechus manatus latirostris)(figure 1). Here are some interesting results of this paper.
Detecting positively selected protein-coding genes
In order to study the molecular mechanism of convergence evolution, firstly, they focused on detecting positive selected protein-coding genes in all three orders; Branch-site likelihood ratio test is a powerful polygenetic method to detect relatively ancient selection. This test is useful for identifying positive selection along prespecified lineages that affects only a few sites in the protein. Applying branch-site likelihood ratio method, they totally tested a series of four different branches. One on the combined marine mammal branches and one on each of the individual branches leading to manatee, walrus and the order containing dolphin and killer whale (see the branches colored red in Fig. 1). They identified 191 genes under positive selection across the combined marine mammal branches, 5 after conservatively correcting for multiple testing.
Identifying Convergent amino acid substitutions in positively selected genes
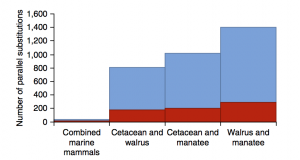
Secondly, they focused on identifying convergent amino acid substitutions encoded within positive selected genes found in the first part. They found such parallel nonsynonymous changes in coding genes mapping to the same amino acid site in more than one marine mammal lineage were widespread across the genome. In a word, they identified 44 parallel nonsynonymous amino acid substitutions occurred along all 3 marine mammal lineages. To specifically, they found 15 of the 44 identical nonsynonymous amino acid substitutionsin all 3 marine mammal lineages encoded within genes evolving under positive selection in at least one lineage; 8 of these genes were inferred to have evolved under positive selection in the test including all 3 marine mammal lineages (Fig. 2 and Table 1).
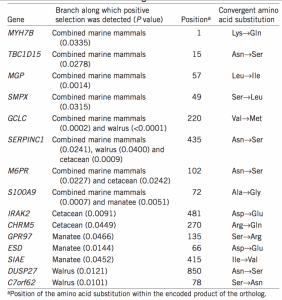
Is phenotype associated with genotype identified in this study? Indeed, they found several of the 15 genes under positive selection have known functional associations that suggest a role in the convergent phenotypic evolution of the marine mammal lineages. For example, S100A9 and MGP encode calcium-binding proteins that have a role in bone formation, SMPX has a role in hearing and inner ear formation16, C7orf62 has known links to hyperthyroidism17, MYH7B has a role in the formation of cardiac muscle18 and SERPINC1 regulates blood coagulation19. These genes could therefore be linked to convergent phenotypic traits such as changes in bone density (S100A9 and MGP), which is high in shallow-diving species such as the manatee and walrus to overcome neutral buoyancy but low in deep-diving cetacean species that collapse their lungs to overcome neutral buoyancy.
For me, the most interesting result they found is an unexpectedly high level of convergence along the combined branches of the terrestrial sister taxa (cow, dog and elephant) to the marine mammals, for which there is no obvious phenotypic convergence. This finding suggests that the options for both adaptive and neutral substitutions in many genes may be limited, possibly because substitutions at alternative sites have pleiotropic and deleterious effects.
Summary
This paper nicely showed that convergent amino acid substitutions were widespread throughout the genome and that a subset of these substitutions were in genes evolving under positive selection and putatively associated with a marine phenotype. However, the authors also found higher levels of convergent amino acid substitutions in a control set of terrestrial sister taxa to the marine mammals. These results suggest that, whereas convergent molecular evolution is relatively common, adaptive molecular convergence linked to phenotypic convergence is comparatively rare.
Foote, A., Liu, Y., Thomas, G., Vina?, T., Alföldi, J., Deng, J., Dugan, S., van Elk, C., Hunter, M., Joshi, V., Khan, Z., Kovar, C., Lee, S., Lindblad-Toh, K., Mancia, A., Nielsen, R., Qin, X., Qu, J., Raney, B., Vijay, N., Wolf, J., Hahn, M., Muzny, D., Worley, K., Gilbert, M., & Gibbs, R. (2015). Convergent evolution of the genomes of marine mammals Nature Genetics, 47 (3), 272-275 DOI: 10.1038/ng.3198
