The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons
About 450 mya bony vertebrates radiated into Lobe-finned fish, from which tetrapods appeared later, and Ray-finned fish, which include Teleost (Fig.1). Nowadays they make up to 96 percent of all fish in the planet. Among the latter some species such as zebrafish (Dario renio) and medaka (Oryzias latipes) are used as model organisms in biomedical research in order to try to understand which is the genetic basis of certain human diseases. However, the transferability between the models is difficult given the phylogenetic distance between tetrapods (humans) and Ray-finned fish. For this reason, the authors decided to sequence the genome of the Spotted Gar (Lepisosteus oculatos), that can act as a bridge as it split off from the teleosts before the TGN (Teleost Genome Duplication). During vertebrate evolution two other genome duplications happened in the vertebrate linage: VGD1 and VGD2.
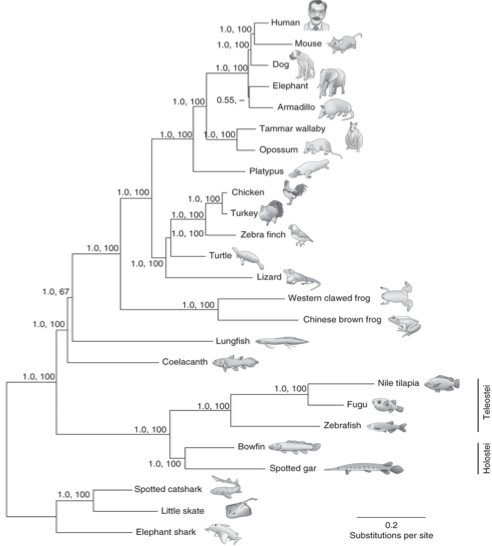
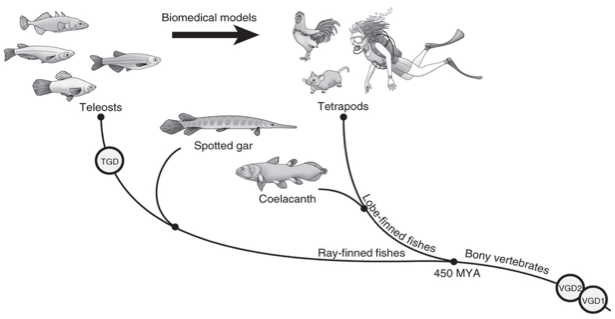 Fig1: Spotted gar is a ray-finned fish that diverged from teleost fishes before the TGD. Gar connects teleosts to lobe-finned vertebrates, such as coelacanth, and tetrapods, including human, by clarifying evolution after the two earlier rounds of vertebrate genome duplication (VGD1 and VGD2) that occurred before the divergence of ray-finned and lobe-finned fishes 450 million years ago (MYA)
Fig1: Spotted gar is a ray-finned fish that diverged from teleost fishes before the TGD. Gar connects teleosts to lobe-finned vertebrates, such as coelacanth, and tetrapods, including human, by clarifying evolution after the two earlier rounds of vertebrate genome duplication (VGD1 and VGD2) that occurred before the divergence of ray-finned and lobe-finned fishes 450 million years ago (MYA)
Genes duplicates derived from the TGN are called Ohnologs. They were named by after Susumu Ohno, who showed in his work genome duplication may play an important role in evolution. The resulting paralogs (a special case of homology when duplicate genes or regions are in the same genome) are associated with development, signaling and gene regulation [2 sentences edited by Marc Robinson-Rechavi]. In addition ohnologs, which amount to about 20 to 35% of genes in the human genome, are frequently implicated in cancer and genetic diseases. Evolution acts on these duplicates and usually they can evolve in three different ways. Mechanisms that lead to preservation of duplicates are sub functionalization (partitioning of ancestral gene functions on the duplicates), neofunctionalization (assigning a novel function to one of the duplicates) and dosage selection (preserving genes to maintain dosage balance between interconnected components). Therefore the most likely outcome is non-functionalization of one duplicate genes due to the lack of selective constraint on preserving both. Because of the asymmetric evolution of ohnologs, TGD, and the speed at which the genome of teleost has evolved, connecting teleost sequences to human sequences can be challenging.
The authors thought, however, that the genome of the Gar can solve these problems due to its slow genetic evolution. Using this “Gar Bridge” allows to clarify the evolution of orthologs (genes in different species that evolved from a common ancestral gene by speciation) in humans such as: (i) Hox and Parahox genes, involved in the formation of body segments during embryogenesis; (ii) The SCPP genes (Calcium binding phosphoproteins), involved in the mineralization of tissues; (iii) miRNA genes, small non-coding RNA molecules that function in RNA silencing and post-transcriptional regulation of gene expression; (iv) CNEs (Conserved Non-coding Elements), regulatory sequences than in previous comparisons between tetrapod and teleost have never appeared. Finally, by the use of transcriptome data they tried to quantify the sum of expression domains and the levels of expression of the TGD-duplicate genes to figure out how these genes evolved.
Genome assembly and annotation
The authors sequenced the genome of one adult female gar to 90x coverage using Illumina technology. By anchoring a scaffold to a meiotic map they captured 94% of assembled bases in 29 linkage groups (LGs). Next, they constructed a gene set composed of 21,433 high confidence protein-coding genes and discovered that 20% of the genome is repetitive with Transposable Elements (TE) that are found in both teleost and lobe-finned fishes. Thanks to this they could clarify the phylogenetic origins of the TE.
The Gar lineage evolved slowly
The authors have made a Bayesian phylogenetic analysis using 243 one-to-one orthologs from 25 jawed vertebrates (Fig.2). Thanks to an evolutionary rate analysis, they showed that the proteins of the sister group of Holostei have evolved more slowly than those of the other vertebrates included in the analysis. These results suggest that the TGD maybe played a role in the rapid evolution of Teleost. The latter is confirmed by the greater branch lengths of the three teleost species used as outgroup.
Fig2: Bayesian phylogeny inferred from 243 proteins with a one-to-one orthology ratio from 25 jawed (gnathostome) vertebrates using PhyloBayes under the CAT + GTR + ?4 model with rooting on cartilaginous fishes. Node support is shown as posterior probability (first number at each node) and bootstrap support from maximum-likelihood analysis (second number at each node).
Gar inform the evolution of bony vertebrate karyotypes
The karyotype of Gar (n2=58), which is composed of micro- and macro-chromosomes, was aligned to those of human, chicken and medaka, a teleost fish. Microchromosomes are present in a wide range of vertebrate classes but not in mammals and teleost. Probably they are the product of an evolutionary process that minimizes the DNA content (mostly through the number of repeats) and maximizes the recombination rate of them. The authors chose the Gar because its genome is the first that does not belong to teleost or lobe finned fish. They could demonstrate a high degree of one-to-one synteny (co-localization of genetic loci on the same chromosome) comparing gar to the chicken genome. This adds support to the hypothesis that the bony ancestor possessed both micro and macro chromosomes. They explain the absence of microchromosomes in teleost by fusion processes that occurred after the divergence from Gar followed by the TGD. In fact, if you look at the comparisons made between Gar and Medaka chromosomes, the synteny relationship is one-to-two meaning that the chromosome sequences are conserved, but are now located on different chromosomes. This confirms that after the fusion and the TGD, teleostei’s chromosomes where subjected to rearrangements and rediploidization and that the radiation of Holostei sister group happened before the genome duplication (Fig.3).
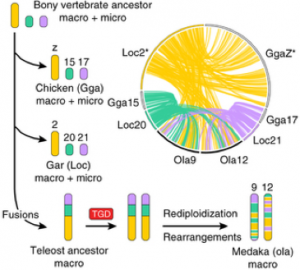 Fig.3: Gar-chicken-medaka comparisons illuminate the karyotype evolution leading to modern teleosts. The genome of the bony vertebrate ancestor contained both macro- and microchromosomes, some of which remain largely conserved in chicken and gar, for example, macrochromosome Loc2-GgaZ and microchromosomes Loc20-Gga15 and Loc21-Gga17. All three chromosomes possess double-conserved synteny with medaka chromosomes Ola9 and Ola12, which is explained by chromosome fusion in the lineage leading to teleosts after divergence from gar, followed by TGD duplication of the fusion chromosome and subsequent intrachromosomal rearrangements and rediploidization.
Fig.3: Gar-chicken-medaka comparisons illuminate the karyotype evolution leading to modern teleosts. The genome of the bony vertebrate ancestor contained both macro- and microchromosomes, some of which remain largely conserved in chicken and gar, for example, macrochromosome Loc2-GgaZ and microchromosomes Loc20-Gga15 and Loc21-Gga17. All three chromosomes possess double-conserved synteny with medaka chromosomes Ola9 and Ola12, which is explained by chromosome fusion in the lineage leading to teleosts after divergence from gar, followed by TGD duplication of the fusion chromosome and subsequent intrachromosomal rearrangements and rediploidization.
Gar clarifies vertebrate gene family evolution
Molecular and physiological mechanisms are shared between vertebrates and this allows to highlight the different types of evolution to which genes were subjected. Despite this after a genome duplication is possible that some ohnologs lineages went lost. The analysis of gar genome allowed to find ancestral genes belonging to VGD1 VGD2 and to clarify the functions of some gene families. For instance, they analyzed the hox family and were able to identify four clusters The number of hox genes that it possesses is greater compared to the ones of tetrapod and teleost. The latter in fact lack some hox orthologs, highlighting that were lost independently in the two groups. The hox genes are very important during embryonic development and intuitively one would think that these have to be more preserved than others. Surprisingly, in my opinion, this study reveal that the teleost, instead of 82 expected Hox cluster genes, have fewer than 50 indicating a massive gene loss after the TGD. The same results were obtained by analyzing circadian clocks, specifically opsin; the MHC’s family; the immunoglobulin genes; the Toll-like receptors. All these genes have shown that gar’s genome can act as a bridge between teleosts and tetrapods, as it possesses characteristics of both.
Gar uncover evolution of vertebrate mineralized tissues
The authors chose this class of proteins because they are preserved for almost all vertebrates. In gar they have an important role as the epidermis is composed of ganoid scales and then formed by ganoin, an “ancestor” of the enamel. However, the evolution of the Scpp (Secretory Calcium-binding Phosphoproteins) was not clear. Gar contain the largest gene number of Scpp, 35, and thanks to this big gene repertory made possible to identify orthologs which with a teleost-tetrapod comparison was not possible to find. The Ambn, Enam and Amel genes, respectively encode ameloblastin, aenamelin and amelogenin. They had been found in the lobe finned fish but not in teleost. These are, however, present in the transcriptome of gar and showing sequence similarity with zebrafish Scpp genes. This suggests that teleost may have different orthologs and that the common ancestor of bony vertebrates had a rich repertoire of Spcc genes. On one hand gar has kept it on the other hand teleosts and tetrapods suffered a loss of subsets of these genes.
Gar connects vertebrate microRNAomes
miRNA is a small non-coding RNA molecule (containing about 22 nucleotides) that functions in RNA silencing and post-transcriptional regulation of gene expression. This gene class has suffered the same evolutionary fate of others mentioned previously. Some sequences have become tetrapod or teleost-specific. The gar genome enabled to identify 107 families. In my opinion the authors did an interesting discover: TGD did not lead to the miRNA loss in teleost. Indeed, the retention rate is higher compared to some protein coding genes, shading new light to the hypothesis that “miRna genes are likely to be retained after a duplication owing their incorporation into multiple gene regulatory networks”. This is evidence of how very often we focus on the evolution of coding sequences of DNA when regulatory mechanisms and non-coding sequences seem to have greater importance.
Gar highlights hidden orthology of cis-regulatory elements
Conserved non-coding element (CNE) are non-coding regions of the genome identified by conventional alignment of genomic sequences from two or more species.
These regions are widely studied because it is unclear the role they play. However, are often considered as cis-acting regulatory sequence (acting on the same molecule of DNA that they regulate). The authors analyzed the evolution of these sequences close to developmental Hox and Parahox genes considering that, during embryonic development, gene expression must be controlled precisely both spatially and temporally. This control is brought about, in large part, by the combinatorial interaction of specific transcription factors with cis-regulatory modules. They chose CNS65, a limb enhancer, because in previous alignments its sequence has been shown to be conserved in humans and chicken but not in teleost. Again using gar CNS65 was possible to find an ortholog in zebrafish. They tested if this cryptic CNS65 enhancer preserves the ancestral function by generate transgenic zebrafish and mice embryos. What they discovered is that the ancestral function was also maintained in zebrafish but with different spatial dynamics. Using mouse embryos, gar CNS65 drives expression of forelimbs and hind limbs in the early stages of development and just later its function is restricted to the distal portion. In zebrafish CNS65 it is only active in the development of the forelimbs (Fig.4).
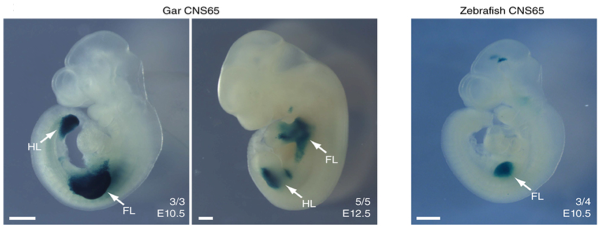 Fig.4: Gar CNS65 drives expression throughout the early mouse forelimbs and hindlimbs (arrows) at stage E10.5 (left). At later stages (E12.5), gar CNS65 activity is restricted to the proximal portion of the limb and is absent in developing digits (middle). Zebrafish CNS65 drives reporter expression in developing mouse limbs at E10.5 but only in forelimbs (right).
Fig.4: Gar CNS65 drives expression throughout the early mouse forelimbs and hindlimbs (arrows) at stage E10.5 (left). At later stages (E12.5), gar CNS65 activity is restricted to the proximal portion of the limb and is absent in developing digits (middle). Zebrafish CNS65 drives reporter expression in developing mouse limbs at E10.5 but only in forelimbs (right).
This is an example of partial loss of the original function, a mechanism that during evolution is more frequent than the gaining of a new function. Besides CNS65 they had found 108 other limb-enhancer in common with humans, compared to 81 that had been found previously with the teleost alignment confirming the presence of hidden orthology (Fig.5).
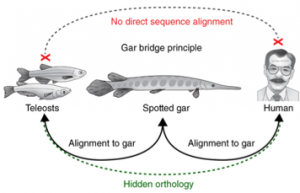 Fig.5: The gar bridge principle of vertebrate CNE connectivity from human through gar to teleosts. Hidden orthology is uncovered for elements that do not directly align between human and teleosts but become evident when first aligning tetrapod genomes to gar, and then aligning gar and teleost genomes
Fig.5: The gar bridge principle of vertebrate CNE connectivity from human through gar to teleosts. Hidden orthology is uncovered for elements that do not directly align between human and teleosts but become evident when first aligning tetrapod genomes to gar, and then aligning gar and teleost genomes
This shows that the latter have suffered the loss of a great number of limb enhancer. In the future, gar will be the ideal candidate to study the limb-to-fin transition.
Gar illuminate gene expression evolution following the TGD
Initially I spoke of evolutionary path that ohnologs (paralog) genes may have after the duplication of the genome. Here the authors were able to get two very clear, I think also very rare, examples as they evolved. The gene slc1a3 went to a neo-functionalization. In gar is expressed only in brain, bone and testis while in medaka, that was chosen by the authors as the representative of the teleost, a ohnolog is mainly expressed in the brain and the other in the liver (Fig.6.c). Completely different fate hit the gpr22 gene that has undergone sub-functionalization. In gar is expressed in the brain and in the heart while in the medaka one ohnolog is expressed in the brain and the other in the heart (Fig.6.d).
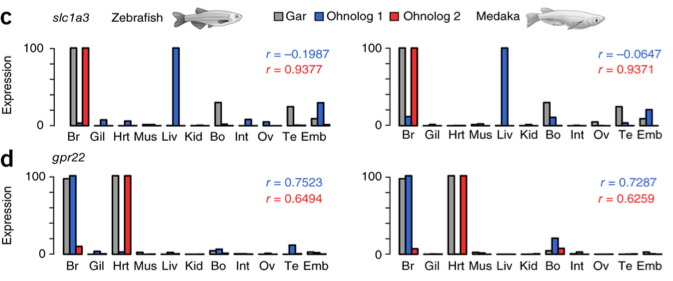 Fig.6: (c) Neofunctionalized ohnologs for slc1a3 showing new expression in liver. (d) Subfunctionalized TGD orthologs of gpr22 with one expressed in brain as in gar and the other expressed in heart as in gar. In c and d, the r values denote the correlation of the expression profile of each ohnolog with the gar pattern.
Fig.6: (c) Neofunctionalized ohnologs for slc1a3 showing new expression in liver. (d) Subfunctionalized TGD orthologs of gpr22 with one expressed in brain as in gar and the other expressed in heart as in gar. In c and d, the r values denote the correlation of the expression profile of each ohnolog with the gar pattern.
This second mechanism is what you would expect with more chances: an ancestral gene sub-function tends to be partitioned between the TGD-derived paralogs. The authors have also seen that the same mechanism occurs regarding the level of gene expression where a ohnologs pair tends to evolve the same level of expression of the pre-duplication gene.
Conclusions
The “Gar-bridges” led to the identification of many ortholog and paralog genes and clarify their fate during evolution. Previously the lack of direct connection between teleost and tetrapod genomes often lead to the wrong use of the word “innovation” on one group or the other. I think that this work is an excellent starting point to connect the evolution of genetic, developmental and physiological mechanisms that made the human genome evolve to its present state. To fully understand the differences between human and model organisms used in biomedicine it is crucial to create very powerful and close-to-reality models. For these reasons, this path should not stop here because the gar is only one species of Holostei – which is composed of nine species and two orders. The study of their genome and also that of other so-called “primitive” fish can help to shine more light on even the striking points that have emerged from this study. Perhaps the outcome of other comparative studies can give even more emphasis to these results or maybe provide answers that may now be counterintuitive.
References
Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi V, Lee AP, Nakamura T, Chalopin D, Fan S, Wcisel D, Cañestro C, Sydes J, Beaudry FE, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SM, Aken B, Yandell M, Schneider I, Yoder JA, Volff JN, Meyer A, Amemiya CT, Venkatesh B, Holland PW, Guiguen Y, Bobe J, Shubin NH, Di Palma F, Alföldi J, Lindblad-Toh K, & Postlethwait JH (2016). The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nature genetics, 48 (4), 427-37 PMID: 26950095