Convergent evolution is defined by the independent evolution of similar traits in different lineages, in order to adapt to similar environmental conditions. Examples of this phenomenon include adaptations to altitude in humans, independent evolution of flight in birds and bats or the multiple evolution of C4 carbon fixation in plants. Yet, the molecular bases of convergent evolution are often lacking. In this paper, authors shed light on the genomic basis of a classical example of convergent evolution, the adaptation to marine life.
Mammals evolved multiple independent times to inhabit the marine environment (Fig 1). Species from three different clades share similar phenotypic adaptations involved in communication, locomotion, thermal regulation, buoyancy… Cetaceans (whales and dolphins) and sirenians (manatees) emerged during the Eocene while pinnipeds (walruses) appeared during Miocene. In this paper, Foote and colleagues investigated the convergent evolution of the genomes of marine mammals at two levels. First, they sought for protein coding genes evolving under positive selection across the three lineages. Second, they studied the convergence of amino acid substitutions within these positively selected genes.
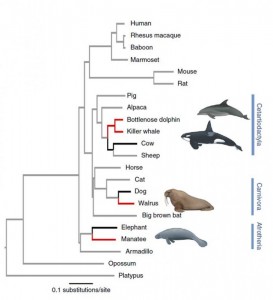
Detection of protein coding genes under positive selection
The authors performed de novo assembly of the genomes of killer whale, manatee and walrus and completed the genome of the bottlenose dolphin using Roche 454 and Illumina HiSeq sequencing. They predicted a set of 16 878 orthologous genes common to the four marine mammal and six terrestrial mammals distributed along the three mammals clades considered here. After filtering, they retained more than 10 000 orthologous genes for each of the four marine mammal species. They identified protein coding genes under positive selection by applying branch-site likelihood ratio test on the three marine mammal lineages alone and combined. Briefly, this test is based on the nonsynonymous to synonymous substitutions rate ratio (dN/dS). The model allows this ratio to vary among sites and among lineages and is then implemented in a likelihood framework. Details of the methods can be found here. They identified 191 genes under positive selection across the combined marine mammal branches, 5 after conservatively correcting for multiple testing.
Convergent amino acid substitutions in marine mammals
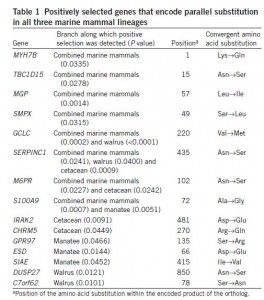
Numerous parallel nonsynonymous substitutions were found in coding genes in pairs of marine mammals (Fig 2). Among them, 44 were common to the three marine mammal lineages with 15 encoded within genes evolving under positive selection (Table 1). This pattern remains even after taking into account hypermutable CpG sites which could bias their results with false positives. Although the precise effect of these parallel substitutions cannot be determined, the functions of the genes that encode these substitutions suggest a role in the convergent phenotypic evolution of mammal for marine life (Supplementary Table 8). Because the three marine mammals lineages considered in this study are relatively distant, these parallel substitutions are likely due to independent de novo mutations in these species, and thus provide strong evidences for convergent molecular evolution.
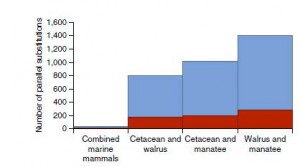
However, a high level of molecular convergence was also found in the terrestrial sister taxa despite no phenotypic convergence. It suggests that parallel substitutions do not commonly result in phenotypic convergence, perhaps because sites that can evolve are limited within genes. Therefore, the molecular changes underlying convergent adaptation are likely to involve other mechanisms than parallel nonsynonymous substitutions.
To conclude, this paper showed that marine mammal evolution partially result from parallel nonsynonymous substitutions. However, it also indicates that convergent molecular evolution seems widespread but do not always result in phenotypic convergence. It suggests that other molecular mechanisms, like convergence of pathways, might be responsible for phenotypic convergence.
Personal comments
This paper reveals the relative importance of convergent molecular evolution in phenotypic convergence. The authors based their study on an important dataset that allows powerful analyses like the branch-site likelihood test. However, they discuss the fact that they found molecular evolution convergence in terrestrial taxa late in the paper, despite this section puts in perspective the importance of their previous findings. In addition, the supplementary materials are not complete and could be clearer. For instance, where are the Table 3, 7 and from 9 to 14? What are the 14 species in the supplementary figure 2? Why are there five pairs of marine mammal in this figure while we should expect more? In addition, this figure illustrates that marine mammals do not show higher convergent molecular evolution than the other species. I think it is an important result that should be in the main text.
Foote, A., Liu, Y., Thomas, G., Vina?, T., Alföldi, J., Deng, J., Dugan, S., van Elk, C., Hunter, M., Joshi, V., Khan, Z., Kovar, C., Lee, S., Lindblad-Toh, K., Mancia, A., Nielsen, R., Qin, X., Qu, J., Raney, B., Vijay, N., Wolf, J., Hahn, M., Muzny, D., Worley, K., Gilbert, M., & Gibbs, R. (2015). Convergent evolution of the genomes of marine mammals Nature Genetics, 47 (3), 272-275 DOI: 10.1038/ng.3198

sorry for my english . i have an interesting argument about the existence of god and evolution: say that we will see a self replicating robot ( lets say even with dna) on a far planet. do we need to conclude design or a natural process in this case? remember that according to evolution if its made from organic components and have a self replicating system we need to conclude a natural process because it has living traits.