Gene duplications are main contributors of genome evolution, but most of the duplicates are redundant and go through pseudogenization. There are several mechanisms proposed to explain how young duplicates survive long-term and escape from being degraded. Among these, dosage-balance model likely to explain the importance of shared expression levels of young duplicate genes. An alternative model indicates sub-functionalization (new copies shares the initial functions) or neo-functionalization (new copy gains new function) as the main mechanisms of the survival of new duplicate. However, it is largely unknown the survival of gene duplication in mammals. In this current study, by using RNA-seq profiles of different human and mouse tissues, authors show that sub-functionalization is a slowly evolving and rare event. Most of the young duplicates are shown to have decreased level of expression, thereby providing initial survival and long-term preservation in the genome.
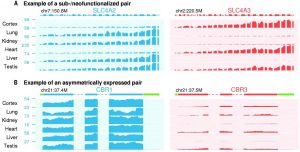
Figure 1 Expression profiles of duplicate genes. Examples of Sub-or Neofunctionalization (A) and asymmetrically expressed gene pairs (B) are shown. In sub-functionalized example, SLC4A2 was shown to be expressed in Lung, Kidney, Liver and Testis, whereas SLC4A3 is expressed in Cortex, Heart and Testis. In asymmetrically expressed gene example, CRB1 is shown to be expressed higher in all tissues that examined.
In order to understand the process of long-term survival after gene duplication, they analyzed RNAseq data of 46 human tissues (from Genotype Tissue Expression, GTEx) and26 mouse tissues. With a computational pipeline (More than %80 coding sequence similarity and more than %50 average sequence similarity), 1444 duplicate gene pairs are identified. These gene pairs are classified as major gene and minor gene, for the higher or lower expression level, respectively. In addition, if a gene pair is at least two-fold higher expressed in minimum one tissue, then it is classified as sub- or neo-functionalized (Figure 1A). Moreover, if a gene pair is expressed more than the other pair in 1/3 of the tissues that examined, it is considered as asymmetrically expressed duplicate (AED) as shown in Figure 1B. Synonymous divergence (ds) was used to estimate divergence time, human-mouse split was shown as 0.45 ds and origin of placental mammals was shown as 0.7 ds.
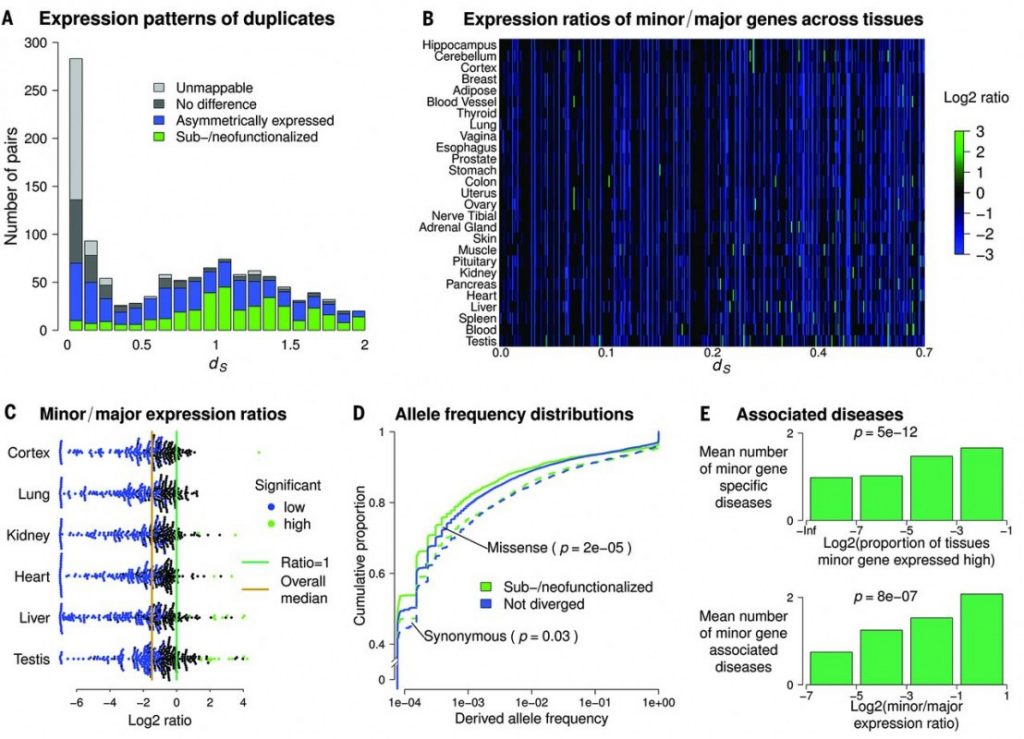 Figure 2 Sub-functionalized or neo-functionalized genes dating back before the emergence of placental mammals.
Figure 2 Sub-functionalized or neo-functionalized genes dating back before the emergence of placental mammals.
Some gene pairs (Mostly of ds < 0.7) are shown to be neo or sub functionalized, yet there are very few examples of neo or sub-functionalization in lately occurred duplication events (Figure 2A-C). In addition, as it is expected that sub-functionalized genes would be under strong selective constraint comparing with non-divergent genes, Kolmogorov-Smirnov test showed that sub-functionalized genes have high fraction rare variants (Figure 2D). Since functionalization would rather give new functions to the gene pairs, authors examined if one of the gene pairs is associated with any disease. There is indeed a correlation that indicating an increase of both minor gene specific disease and minor gene associated disease, when there is a sub-functionalization event (Figure 2E).
The duplicates that are risen within placental mammals, most duplicate pairs are shown to be AEDs other than sub-functionalized and within AEDs, very few minor genes are associated with disease in contrast to what was shown in Figure 2E. All these results indicate that, sub-functionalization is a slowly evolving event, although it was shown that duplicates on different chromosomes have higher rates or neo- or sub-functionalization when it is compared with duplicates that are in tandem arrays. This brings the question, whether separation of the duplicates is a facilitating process for sub-functionalization.
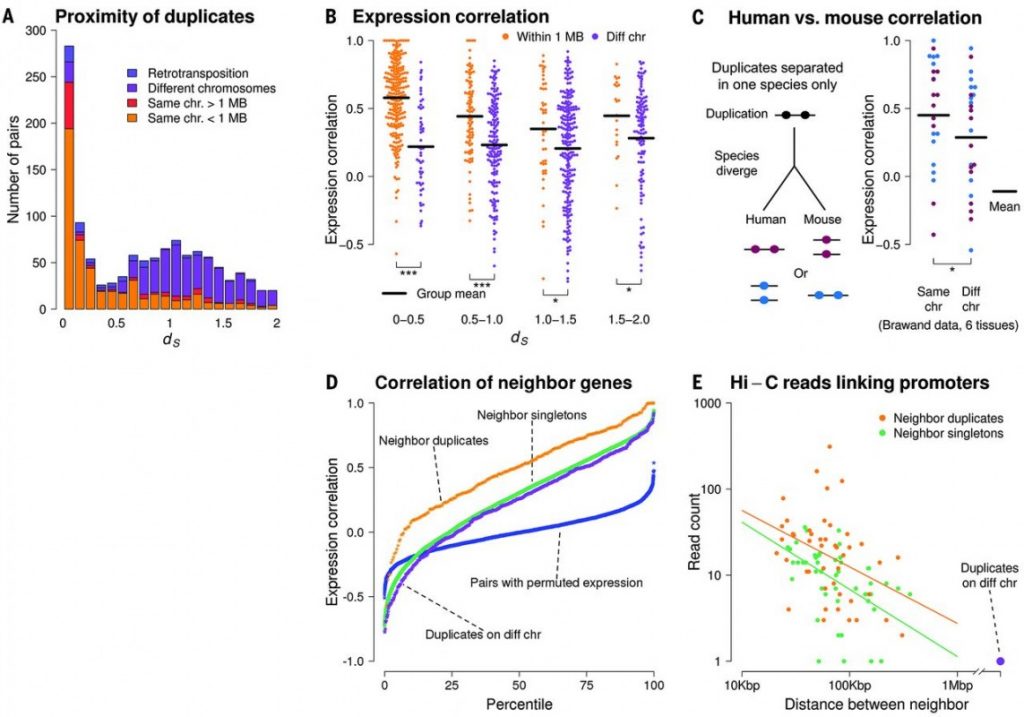 Figure 3 Genomic Location of the duplicates and expression correlation. It is shown that most of the young duplicates are located in same chromosome and are closely located to each other, whereas the older duplicates tend to locate on different chromosomes. Depending on how closely the duplicates locate on the chromosome (both in human and mouse), there is a higher of expression correlation of the duplicates.
Figure 3 Genomic Location of the duplicates and expression correlation. It is shown that most of the young duplicates are located in same chromosome and are closely located to each other, whereas the older duplicates tend to locate on different chromosomes. Depending on how closely the duplicates locate on the chromosome (both in human and mouse), there is a higher of expression correlation of the duplicates.
Supporting this idea, authors indicated that 87% young gene pairs with ds < 0.1 are found in tandem arrays in the same chromosomes (Figure 3A). The rest of the duplicates found on different chromosomes are most likely separated by the result of chromosomal rearrangements and they have diverged expression pattern due to the genomic separation (Figure 3B). It is shown that the more genomic distance of the duplicates increases, the less expression correlation of the duplicates is observed. Notably, it is also shown that duplicates in mouse have a similar correlation with human duplicates, indicating the negative relation between genomic distance and expression correlation is not human specific (Figure 3C). This data supports what was previously shown about the coregulation of closely located genes in the genome and it is once shown in Figure 3D, as neighbor duplicates have higher expression correlation comparing with duplicates on different chromosomes and singletons. In addition, whole-genome chromosome conformation capture (Hi-C) shows that neighboring duplicates have higher connectivity and more promoter-promoter links comparing with neighboring singletons (Figure 3D).
So far, it is shown that expression sub-functionalization is a slowly evolving process and duplicates that are in tandem arrays are mostly coregulated. As an alternative explanation, if dosage sharing is crucial for the preservation of newborn duplicates, it must be shown that there is a shared and lower expression of the duplicates. In order to prove this hypothesis, the authors investigated the human duplicates since human-macaque split with RNA-seq results of six different tissues. It is obvious that, the sum of expression levels of human major and minor duplicate is corresponding to the expression level of macaque singleton ortholog (Figure 4A). This data proves that dosage sharing is a fast evolving event, contributing to the preservation of duplicates in the genome.
 Figure 4 Dosage sharing and multi-step model of how duplicate genes are preserved. Summed expression of human young duplicate is similar to the expression of macaque ortholog.
Figure 4 Dosage sharing and multi-step model of how duplicate genes are preserved. Summed expression of human young duplicate is similar to the expression of macaque ortholog.
Overall, in this current study the mechanism of how duplicated genes are preserved is explained with a multi-step model (Figure 4C). According to the model, after a duplication event happens, expression dosage is shared between two duplicates which was also suggested for whole genome duplications. In this process, there is a tight competition between dosage sharing and mutational degradation of one of the duplicates. After this important step, minor gene of the asymmetrically expressed duplicate can be lost slowly under reduced constraint. In an alternative long-term scenario, chromosomal rearrangements would happen to separate the coregulation of these tandem duplicates and providing different expression pattern and/or protein adaptation which will cause long-term survival of the duplicated genes. To sum up, this study shows that rapid dose sharing is a fundamental first step after the duplication of a gene and it can be followed by a slow evolving subfunctionalization event of the duplicate.
References
Xun Lan and Jonathan K. Pritchard
Science 20 May 2016:
Vol. 352, Issue 6288, pp. 1009-1013
DOI: 10.1126/science.aad8411